
https://ninjani.github.io/
www.biorxiv.org/content/10.1...
🧵👇 (1/n)

Day 2/31
Prompt WEAVE
N-terminal domain of a Fibrion - a building block of silk fiber produced by silkworms.
Pdb: 3UA0
Next prompt is CROWN and I would love your suggestions!

Day 2/31
Prompt WEAVE
N-terminal domain of a Fibrion - a building block of silk fiber produced by silkworms.
Pdb: 3UA0
Next prompt is CROWN and I would love your suggestions!
~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...

~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...
“From bytes to binders: design, score and optimize” #bc2basel #posterprize
“From bytes to binders: design, score and optimize” #bc2basel #posterprize
@sib.swiss @biozentrum.unibas.ch
⬇️⬇️⬇️
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

@sib.swiss @biozentrum.unibas.ch
⬇️⬇️⬇️
www.nature.com/articles/s41...
www.nature.com/articles/s41...
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.
Find out about molecular modelling to systems-level analyses and evolution, and more.
Submit your abstract by 26 Aug ➡️ s.embl.org/csb25-01-bl

Find out about molecular modelling to systems-level analyses and evolution, and more.
Submit your abstract by 26 Aug ➡️ s.embl.org/csb25-01-bl
We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...

We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6

AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
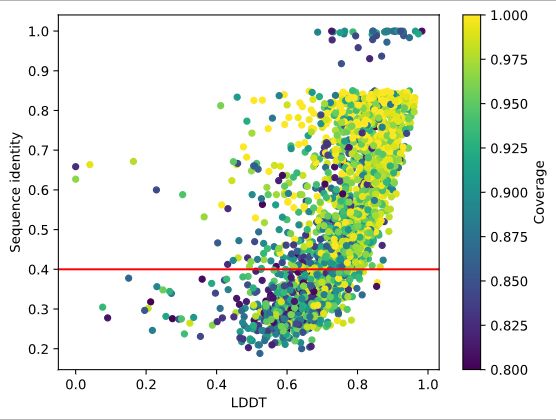
The summer call at @biozentrum.unibas.ch @unibas.ch is open until October 12, 2025.
www.biozentrum.unibas.ch/phd/internat...

The summer call at @biozentrum.unibas.ch @unibas.ch is open until October 12, 2025.
www.biozentrum.unibas.ch/phd/internat...
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

patwalters.github.io/Three-Papers...

patwalters.github.io/Three-Papers...

@torstenschwede.bsky.social @xavier.robin.info
Join us and register your server now!
cameo3d.org
@torstenschwede.bsky.social @xavier.robin.info
Join us and register your server now!
cameo3d.org
Join us and register your server now!
cameo3d.org
Join us and register your server now!
cameo3d.org
If you are excited about combining AI and machine learning with fundamental biology, we would love to hear from you!
https://vib.ai/en/opportunities#/job-description/110906

If you are excited about combining AI and machine learning with fundamental biology, we would love to hear from you!
https://vib.ai/en/opportunities#/job-description/110906

