Leveraging long-read metagenomics (@nanoporetech.com) we identified some of the most prevalent gut phage families that have previously been overlooked in short-read based studies. [1/5]
Read more here: www.biorxiv.org/content/10.6...
Leveraging long-read metagenomics (@nanoporetech.com) we identified some of the most prevalent gut phage families that have previously been overlooked in short-read based studies. [1/5]
Read more here: www.biorxiv.org/content/10.6...
Leveraging long-read metagenomics (@nanoporetech.com) we identified some of the most prevalent gut phage families that have previously been overlooked in short-read based studies. [1/5]
Read more here: www.biorxiv.org/content/10.6...
By integrating a single-cell dispenser into a culturomics workflow, we accelerated the isolation of individual bacterial cells, allowing robust strain-level resolution.

@isaacyueyuan.bsky.social, for this wonderful write-up. I would also want to thank @niranjantw.bsky.social and @jsgounot.bsky.social for their uwavering support and guidance.
By integrating a single-cell dispenser into a culturomics workflow, we accelerated the isolation of individual bacterial cells, allowing robust strain-level resolution.

@isaacyueyuan.bsky.social, for this wonderful write-up. I would also want to thank @niranjantw.bsky.social and @jsgounot.bsky.social for their uwavering support and guidance.
Great to have this work out early in the year:
journals.asm.org/eprint/KHNHX...
We report our experience with the B.SIGHT system from @cytena.bsky.social
Great to have this work out early in the year:
journals.asm.org/eprint/KHNHX...
We report our experience with the B.SIGHT system from @cytena.bsky.social
nature.com/articles/s44...

nature.com/articles/s44...
1) Defined spatial sampling can provide in vivo relevant insights
2) Microbial mRNA is dominant, but human mRNA is also readily detected and quantified
www.nature.com/articles/s41...

1) Defined spatial sampling can provide in vivo relevant insights
2) Microbial mRNA is dominant, but human mRNA is also readily detected and quantified
www.nature.com/articles/s41...
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...

Read all about it here: nature.com/articles/s41...
Great collab with Chew and Hirao lab
x.com/NiranjanTW/s...

Read all about it here: nature.com/articles/s41...
Great collab with Chew and Hirao lab
x.com/NiranjanTW/s...
Full story below👇:
Full story below👇:
@niranjantw.bsky.social www.nature.com/articles/s41...
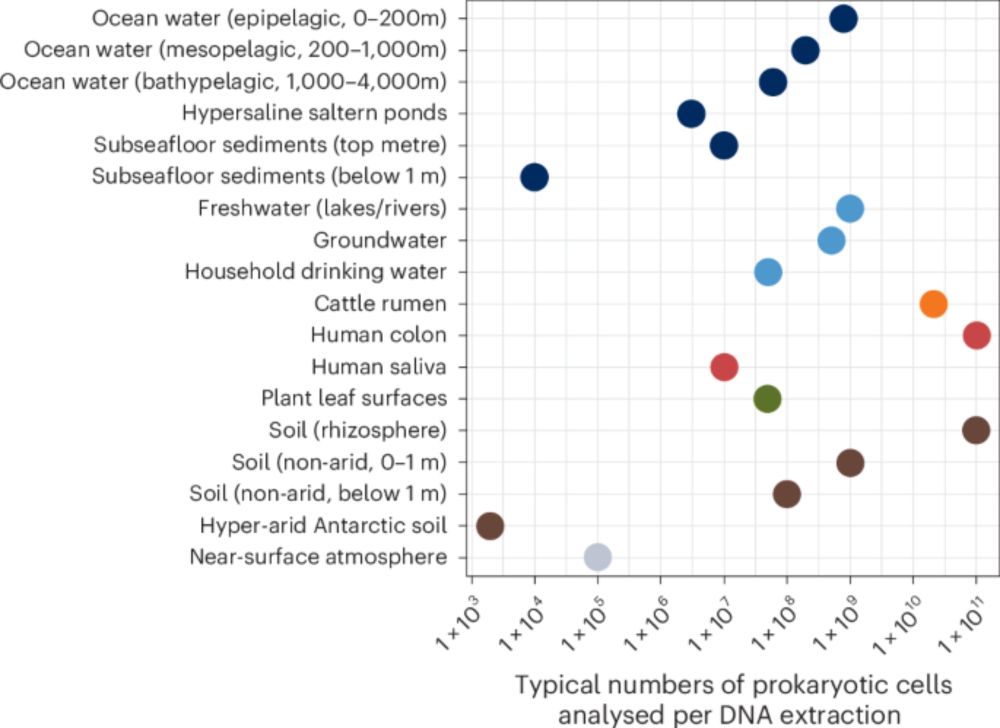
@niranjantw.bsky.social www.nature.com/articles/s41...



