
Bacterial and plasmid evolution 🫧🧬💻

Do more plasmid copies mean faster evolution?
🧵 Dive into the story
www.pnas.org/doi/10.1073/...


We represent plasmids as circles and mutations as dots, resembling an eye, because in this paper we literally 𝑤𝑎𝑡𝑐ℎ plasmids evolve.
‼️Check Paula’s 🧵 and the paper👇
𝗣𝗹𝗮𝘀𝗺𝗶𝗱 𝗺𝘂𝘁𝗮𝘁𝗶𝗼𝗻 𝗿𝗮𝘁𝗲𝘀 𝘀𝗰𝗮𝗹𝗲 𝘄𝗶𝘁𝗵 𝗰𝗼𝗽𝘆 𝗻𝘂𝗺𝗯𝗲𝗿
www.pnas.org/doi/10.1073/...
Do more plasmid copies mean faster evolution?
🧵 Dive into the story
www.pnas.org/doi/10.1073/...

We represent plasmids as circles and mutations as dots, resembling an eye, because in this paper we literally 𝑤𝑎𝑡𝑐ℎ plasmids evolve.
‼️Check Paula’s 🧵 and the paper👇
𝗣𝗹𝗮𝘀𝗺𝗶𝗱 𝗺𝘂𝘁𝗮𝘁𝗶𝗼𝗻 𝗿𝗮𝘁𝗲𝘀 𝘀𝗰𝗮𝗹𝗲 𝘄𝗶𝘁𝗵 𝗰𝗼𝗽𝘆 𝗻𝘂𝗺𝗯𝗲𝗿
www.pnas.org/doi/10.1073/...

We represent plasmids as circles and mutations as dots, resembling an eye, because in this paper we literally 𝑤𝑎𝑡𝑐ℎ plasmids evolve.
‼️Check Paula’s 🧵 and the paper👇
𝗣𝗹𝗮𝘀𝗺𝗶𝗱 𝗺𝘂𝘁𝗮𝘁𝗶𝗼𝗻 𝗿𝗮𝘁𝗲𝘀 𝘀𝗰𝗮𝗹𝗲 𝘄𝗶𝘁𝗵 𝗰𝗼𝗽𝘆 𝗻𝘂𝗺𝗯𝗲𝗿
www.pnas.org/doi/10.1073/...

Do more plasmid copies mean faster evolution?
🧵 Dive into the story
www.pnas.org/doi/10.1073/...

Do more plasmid copies mean faster evolution?
🧵 Dive into the story
www.pnas.org/doi/10.1073/...
Here we explore the conjugative capacity of these mysterious Genomic Islands.
www.biorxiv.org/content/10.6...
Here we explore the conjugative capacity of these mysterious Genomic Islands.
www.biorxiv.org/content/10.6...
Here we explore the conjugative capacity of these mysterious Genomic Islands.
www.biorxiv.org/content/10.6...
The long-term existence of diverse virulent phages within cultures of Escherichia coli and others challenges the virulent–temperate dichotomy and points to non-canonical phage lifestyles.
#MicroSky 🦠 @peterdoug.bsky.social @emggroupucph.bsky.social
www.nature.com/articles/s41...

The long-term existence of diverse virulent phages within cultures of Escherichia coli and others challenges the virulent–temperate dichotomy and points to non-canonical phage lifestyles.
#MicroSky 🦠 @peterdoug.bsky.social @emggroupucph.bsky.social
www.nature.com/articles/s41...
Diverse genomes of lytic phages are found in bacterial assemblies, challenging assumptions about the nature of the lytic lifestyle.
#MicroSky 🦠
www.nature.com/articles/s41...

Diverse genomes of lytic phages are found in bacterial assemblies, challenging assumptions about the nature of the lytic lifestyle.
#MicroSky 🦠
www.nature.com/articles/s41...
Happy holidays, everyone! 🎄✨
Happy holidays, everyone! 🎄✨
@microryc.bsky.social @esgem-sg.bsky.social
@microryc.bsky.social @esgem-sg.bsky.social
Plasmid copy number ≈ 2.5% of chromosome size—consistent across bacterial species!
pmc.ncbi.nlm.nih.gov/articles/PMC... @jerorb.bsky.social
🧪 #microbesky

Plasmid copy number ≈ 2.5% of chromosome size—consistent across bacterial species!
pmc.ncbi.nlm.nih.gov/articles/PMC... @jerorb.bsky.social
🧪 #microbesky

Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
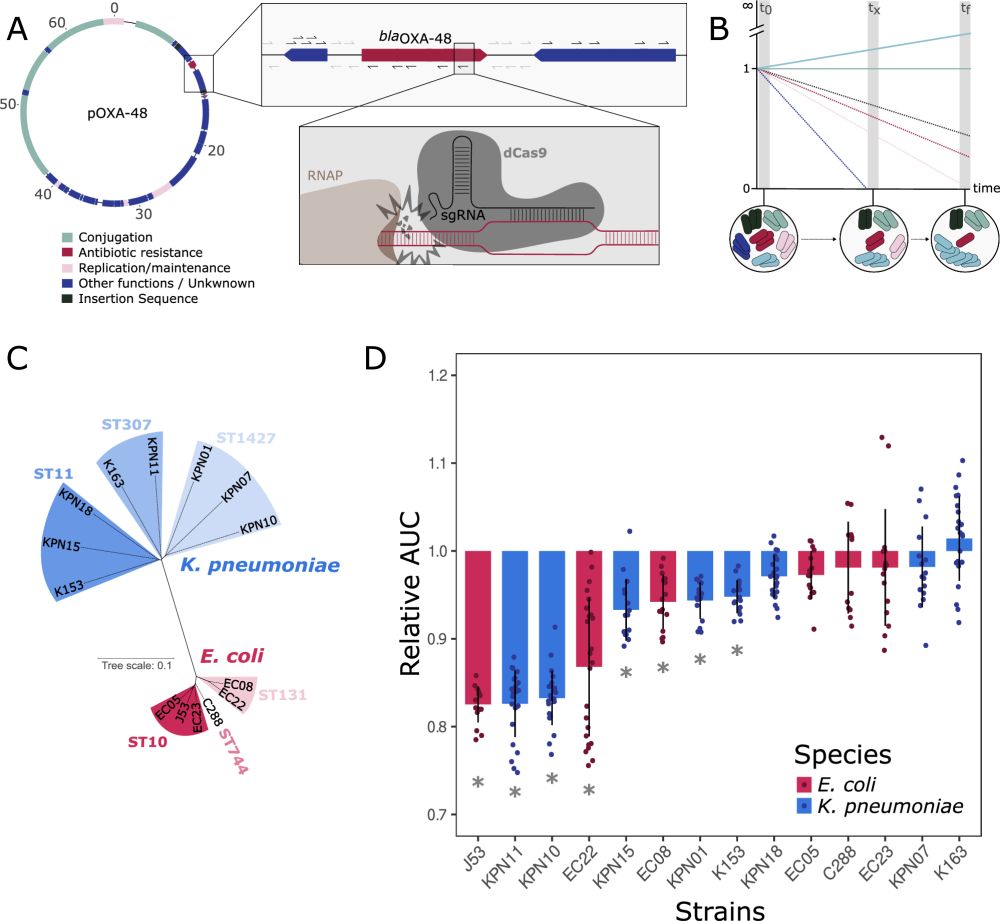
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
Plasmids ✅
Insertion Sequences ✅
AMR Evolution ✅
Microbial Communities ✅
Databases analyses ✅
Mathematical modeling ✅
See the scientific thread below of Jorge Sastre, who has brilliantly led this work with @palomarodera.bsky.social
Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation.
Combining experimental and computational approaches, we unveil how two of the most prevalent bacterial MGE accelerate the evolution of AMR. 🧵👇🏻
www.biorxiv.org/content/10.1...

Plasmids ✅
Insertion Sequences ✅
AMR Evolution ✅
Microbial Communities ✅
Databases analyses ✅
Mathematical modeling ✅
See the scientific thread below of Jorge Sastre, who has brilliantly led this work with @palomarodera.bsky.social
Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation.
Combining experimental and computational approaches, we unveil how two of the most prevalent bacterial MGE accelerate the evolution of AMR. 🧵👇🏻
www.biorxiv.org/content/10.1...

Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation.
Combining experimental and computational approaches, we unveil how two of the most prevalent bacterial MGE accelerate the evolution of AMR. 🧵👇🏻
www.biorxiv.org/content/10.1...
Excludons are pairs of overlapping genes that block each other’s expression (basically, reverse operons).
We built a tool to identify them in bacterial genomes using transcriptomic data, in an awesome collab led by Iñigo Lasa and Álvaro San Martín.
👇
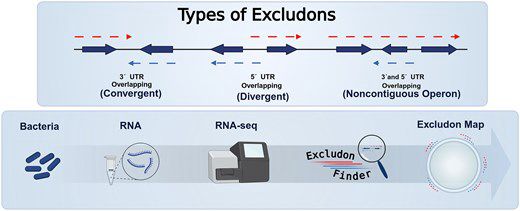
Excludons are pairs of overlapping genes that block each other’s expression (basically, reverse operons).
We built a tool to identify them in bacterial genomes using transcriptomic data, in an awesome collab led by Iñigo Lasa and Álvaro San Martín.
👇


In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇

Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇

Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇

