
neurips.cc/virtual/2025...
arxiv.org/abs/2507.00846

neurips.cc/virtual/2025...
arxiv.org/abs/2507.00846
We show how trajectories of spatial dynamical systems can be modeled in latent space by
--> leveraging IDENTIFIERS.
📚Paper: arxiv.org/abs/2502.12128
💻Code: github.com/ml-jku/LaM-S...
📝Blog: ml-jku.github.io/LaM-SLidE/
1/n

We show how trajectories of spatial dynamical systems can be modeled in latent space by
--> leveraging IDENTIFIERS.
📚Paper: arxiv.org/abs/2502.12128
💻Code: github.com/ml-jku/LaM-S...
📝Blog: ml-jku.github.io/LaM-SLidE/
1/n

The best part was using @iclr-conf.bsky.social as my personal meme account the past two years (first as Senior Program Chair). I hope that future ICLR organizers continue the tradition. 🤪

The best part was using @iclr-conf.bsky.social as my personal meme account the past two years (first as Senior Program Chair). I hope that future ICLR organizers continue the tradition. 🤪
A collaboration between the groups of @martinsteinegger.bsky.social , David Jones and Christine Orengo, we clustered AlphaFold Database and ESMatlas, a whopping 821 million proteins!
We reveal biome-specific groups & over 11k novel domain combinations.

A collaboration between the groups of @martinsteinegger.bsky.social , David Jones and Christine Orengo, we clustered AlphaFold Database and ESMatlas, a whopping 821 million proteins!
We reveal biome-specific groups & over 11k novel domain combinations.
I am very excited to share with you the corporate identity of AITHYRA, the Research Institute for Biomedical AI.
Check out the brand design video (sound on!) to learn more.
**REMINDER**
One week left to apply for Starting PI positions (Life Science and AI/ML). Come join us!
I am very excited to share with you the corporate identity of AITHYRA, the Research Institute for Biomedical AI.
Check out the brand design video (sound on!) to learn more.
**REMINDER**
One week left to apply for Starting PI positions (Life Science and AI/ML). Come join us!
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...



Check out our latest paper on using reinforcement learning for pharmacophore elucidation on protein binding sites. It comes with a google colab notebook for use.
bmcbiol.biomedcentral.com/articles/10....

Check out our latest paper on using reinforcement learning for pharmacophore elucidation on protein binding sites. It comes with a google colab notebook for use.
bmcbiol.biomedcentral.com/articles/10....
Check out each of their papers below:

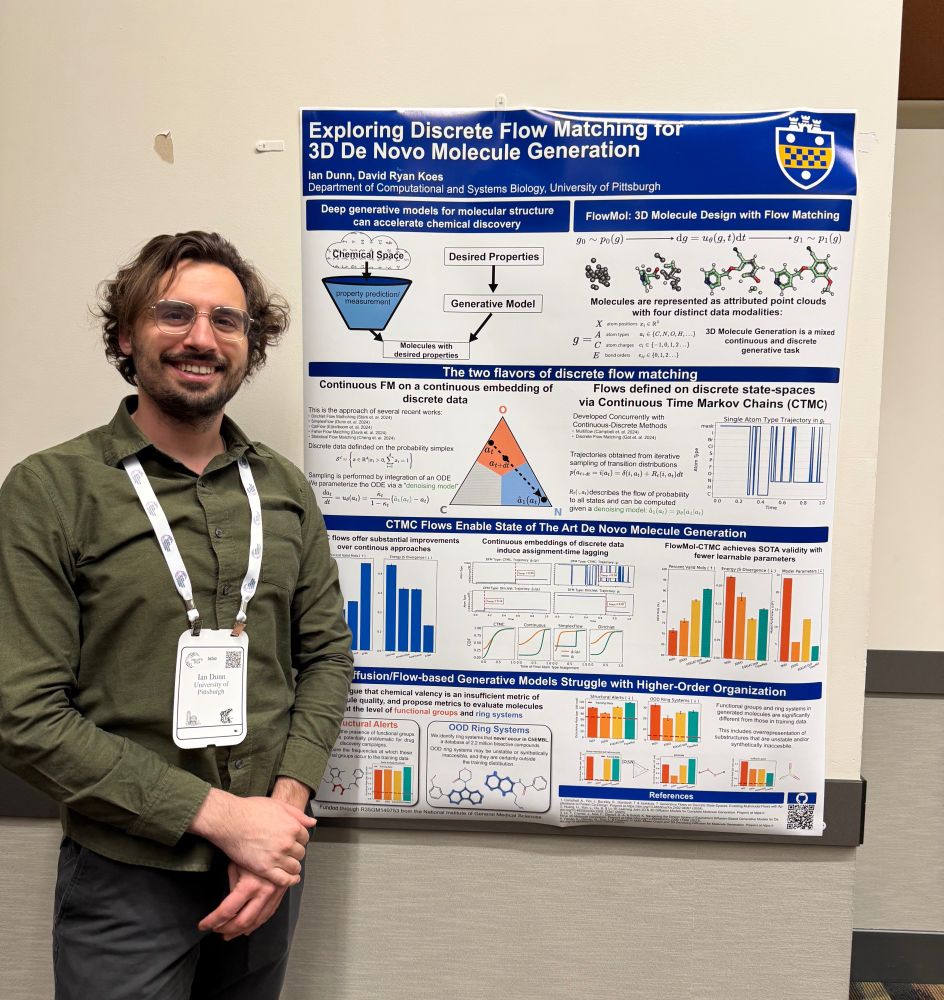
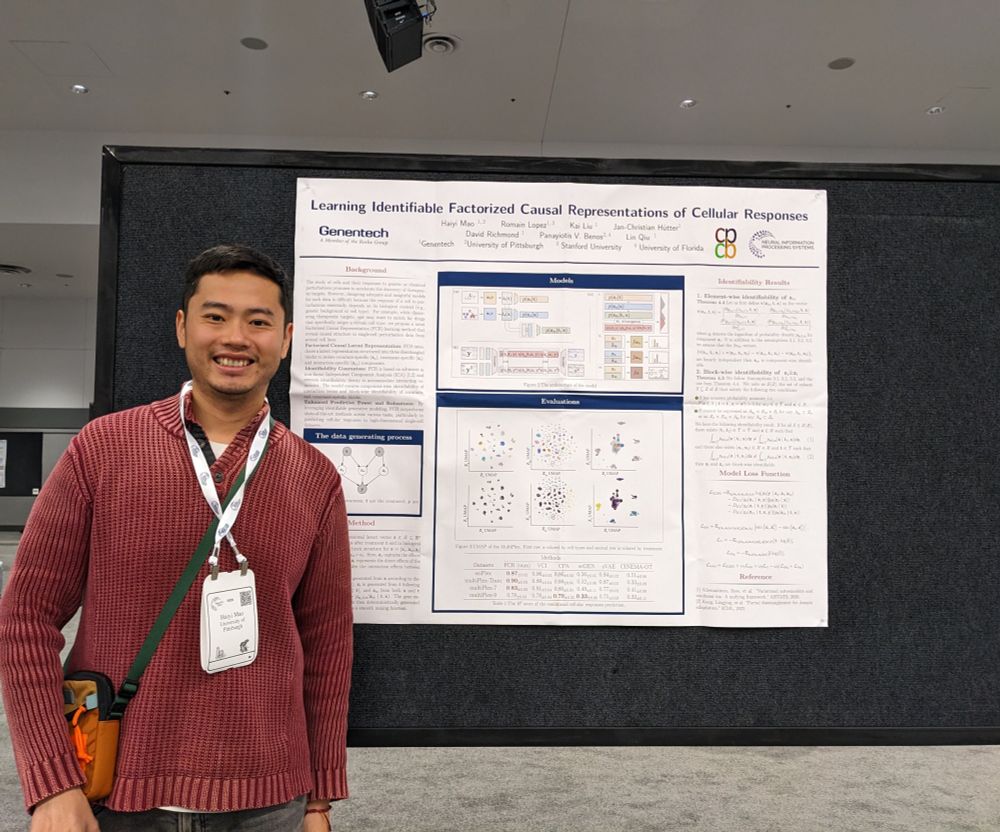
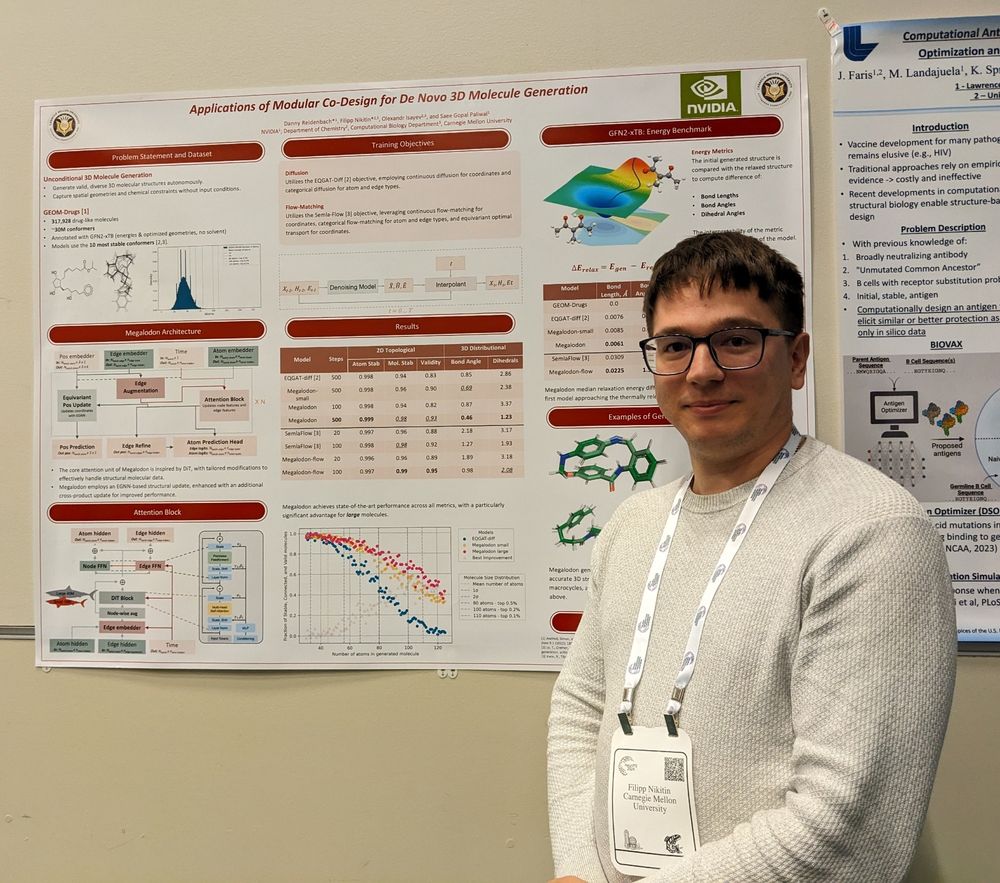
Check out each of their papers below:

Opening this position for just a few days over #NeurIPS2024. Looking especially for Bioinformatics + Structural Biology skills, MD/Stat Mech skills and/or #deeplearning architecture design + engineering skills
aka.ms/ai4science-r...
Opening this position for just a few days over #NeurIPS2024. Looking especially for Bioinformatics + Structural Biology skills, MD/Stat Mech skills and/or #deeplearning architecture design + engineering skills
aka.ms/ai4science-r...
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
For good or for bad, in the end, BLINDED COMMUNITY CHALLENGES are the only fair way to assess methods
Meaning: the proof is in the pudding. The methods who want to be taken seriously should ...
For good or for bad, in the end, BLINDED COMMUNITY CHALLENGES are the only fair way to assess methods
Meaning: the proof is in the pudding. The methods who want to be taken seriously should ...



