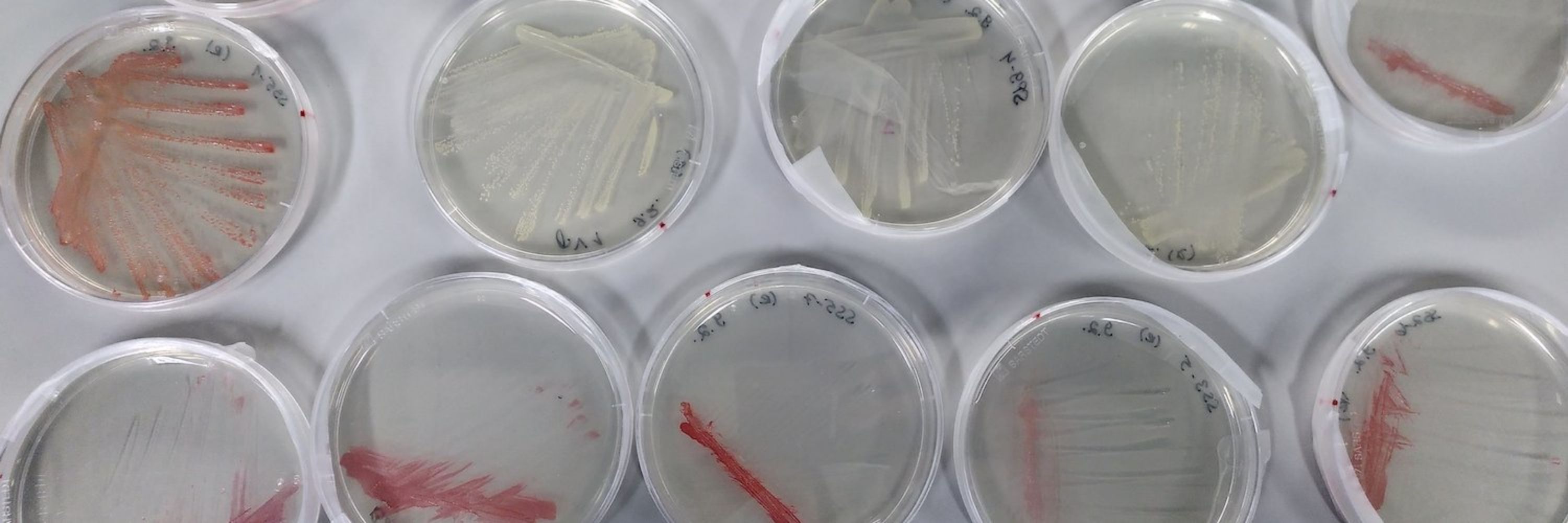

journals.plos.org/plosbiology/...
"The origin of life on Earth remains one of the greatest and most pervasive mysteries in science. We know the story in broad strokes: Around 4 billion years ago, simple chemical compounds gave rise to living cells, which later formed..."
🦠
asm.org/articles/202...
"The origin of life on Earth remains one of the greatest and most pervasive mysteries in science. We know the story in broad strokes: Around 4 billion years ago, simple chemical compounds gave rise to living cells, which later formed..."
🦠
asm.org/articles/202...
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...



@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
A fantastic collaboration with Antoine, with Jovana Kaljevic' initiated the collaboration and drives the project.

Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...

Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...
Structure of a functional archaellum in bacteria of the Chloroflexota phylum
#MicroSky
@archaellum.bsky.social @sshamphavi.bsky.social @loumollat.bsky.social @mariejoest.bsky.social @sgribaldo.bsky.social
www.nature.com/articles/s41...

Structure of a functional archaellum in bacteria of the Chloroflexota phylum
#MicroSky
@archaellum.bsky.social @sshamphavi.bsky.social @loumollat.bsky.social @mariejoest.bsky.social @sgribaldo.bsky.social
www.nature.com/articles/s41...
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...

It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September

Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September
Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵

In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
asm.org/podcasts/twi...
asm.org/podcasts/twi...

"The humble bacterium is still a relevant tool for the study of the underlying mechanisms that are conserved throughout life."
🧪🧫🧬📚
doi.org/10.1093/gene...

"The humble bacterium is still a relevant tool for the study of the underlying mechanisms that are conserved throughout life."
🧪🧫🧬📚
doi.org/10.1093/gene...
🧵 [1/n]
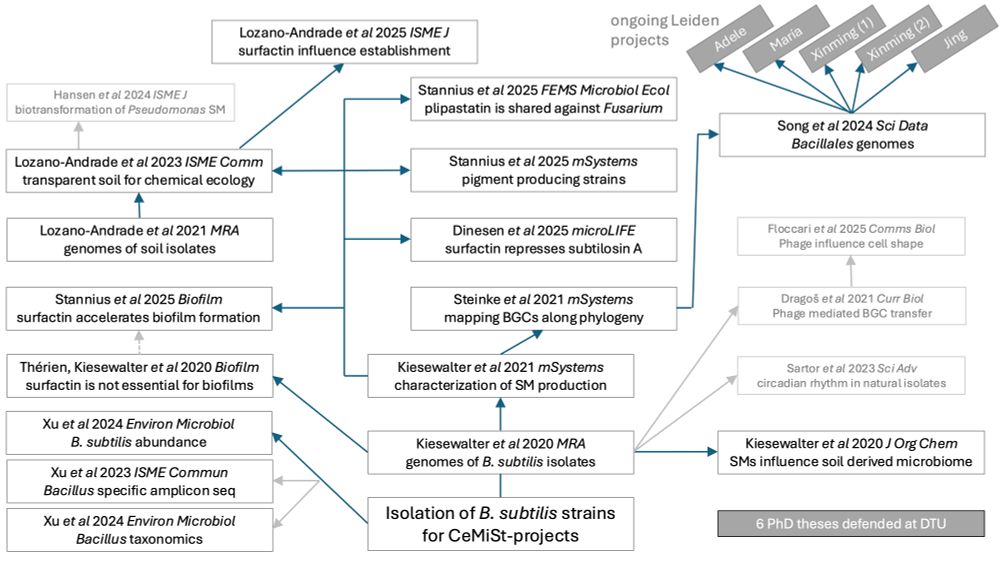

🧵 [1/n]
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12

Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12
Register here: register.oxfordabstracts.com/event/75604?...
Abstract submission: app.oxfordabstracts.com/stages/79029...
Join us for a day of talks, networking and career discussions. Present your work to get fresh new ideas and the chance to win prizes 💸
Featuring @avsecz.bsky.social of Google DeepMind as our keynote speaker⚡️

Register here: register.oxfordabstracts.com/event/75604?...
Abstract submission: app.oxfordabstracts.com/stages/79029...

journals.plos.org/plosbiology/...

journals.plos.org/plosbiology/...
Why should you care? Because it helps shift our view of archaea — from rare extremophiles to not-so-passive bystanders. Some archaea can also fend off bacteria.
Glad to be part of this cool project from @tobiaswarnecke.bsky.social. Opens lots of Questions.
Work by @romainstrock.bsky.social shows that some archaea can kill bacteria by secreting peptidoglycan hydrolases. journals.plos.org/plosbiology/...
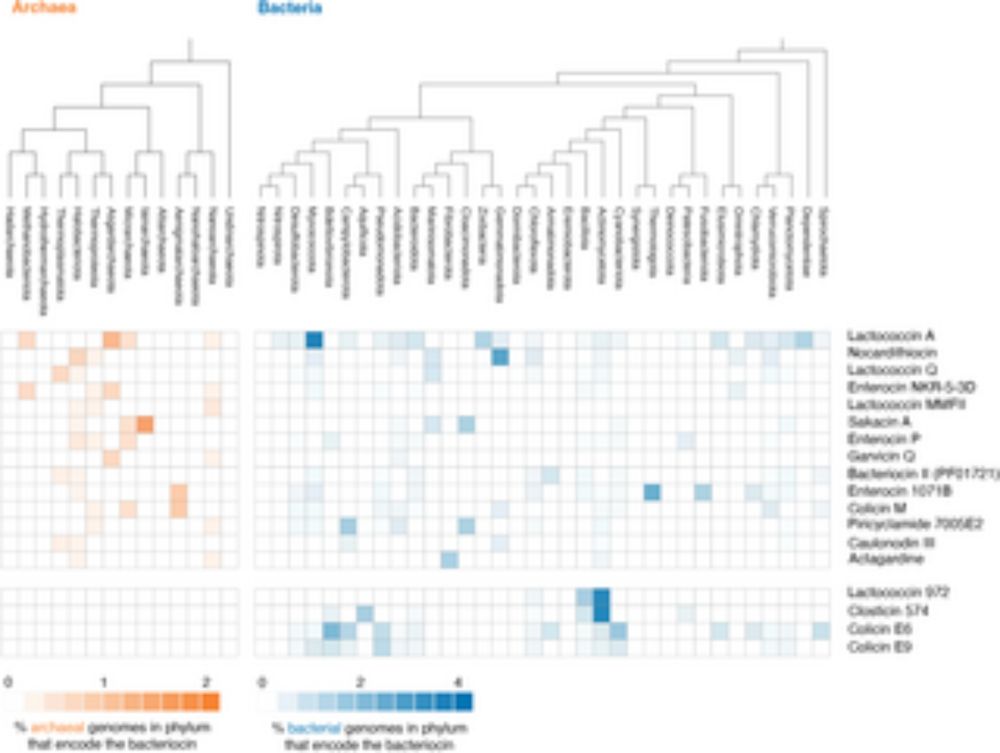
Why should you care? Because it helps shift our view of archaea — from rare extremophiles to not-so-passive bystanders. Some archaea can also fend off bacteria.
Glad to be part of this cool project from @tobiaswarnecke.bsky.social. Opens lots of Questions.
Work by @romainstrock.bsky.social shows that some archaea can kill bacteria by secreting peptidoglycan hydrolases. journals.plos.org/plosbiology/...

Work by @romainstrock.bsky.social shows that some archaea can kill bacteria by secreting peptidoglycan hydrolases. journals.plos.org/plosbiology/...

Work by @romainstrock.bsky.social shows that some archaea can kill bacteria by secreting peptidoglycan hydrolases. journals.plos.org/plosbiology/...
With @tweethinking.bsky.social @sabifo4.bsky.social @ssolo.bsky.social Davide Pisani, Tim Lenton and @phil-donoghue.bsky.social
(1/2)

With @tweethinking.bsky.social @sabifo4.bsky.social @ssolo.bsky.social Davide Pisani, Tim Lenton and @phil-donoghue.bsky.social
(1/2)


