rrwick.github.io/2023/04/19/v...
Now uses qsv instead of xsv, plus some other miscellaneous tweaks/fixes.
In the 2+ years since I wrote this post, I use my tv function a LOT.
rrwick.github.io/2023/04/19/v...
Now uses qsv instead of xsv, plus some other miscellaneous tweaks/fixes.
In the 2+ years since I wrote this post, I use my tv function a LOT.
metaMDBG (@gaetanbenoit.bsky.social) and Myloasm (@jimshaw.bsky.social) have had recent releases, so I updated the benchmarks from the Autocycler paper:
rrwick.github.io/2025/09/23/a...
Both tools improved considerably! Time to update your conda environments 😄
metaMDBG (@gaetanbenoit.bsky.social) and Myloasm (@jimshaw.bsky.social) have had recent releases, so I updated the benchmarks from the Autocycler paper:
rrwick.github.io/2025/09/23/a...
Both tools improved considerably! Time to update your conda environments 😄
www.biorxiv.org/content/10.1...
#Bioinformatics #genomics #assembly #assemblygraphs #software

www.biorxiv.org/content/10.1...
#Bioinformatics #genomics #assembly #assemblygraphs #software
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
I added a new feature to @gbouras13.bsky.social's Pypolca: homopolymer-only polishing. Potentially useful for cross-sample polishing - early test on Cryptosporidium looks promising.
Check it out here:
rrwick.github.io/2025/09/04/h...
I added a new feature to @gbouras13.bsky.social's Pypolca: homopolymer-only polishing. Potentially useful for cross-sample polishing - early test on Cryptosporidium looks promising.
Check it out here:
rrwick.github.io/2025/09/04/h...
www.medrxiv.org/content/10.1...
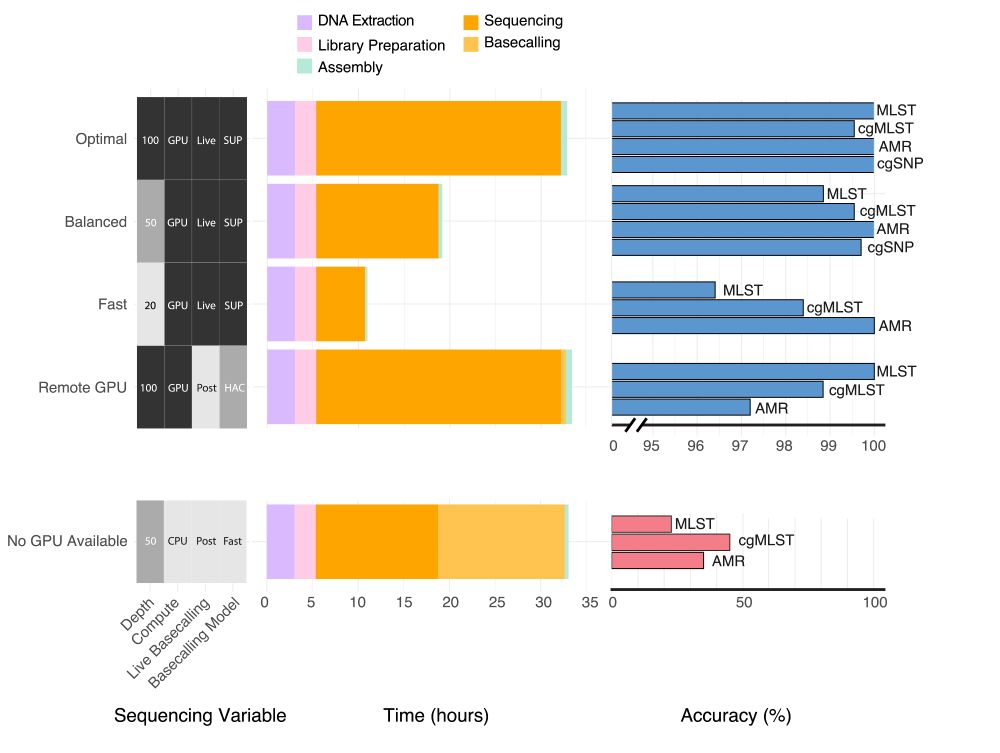
www.medrxiv.org/content/10.1...
github.com/nanoporetech...
It supports using --bacteria when polishing an assembly with v5.2.0 data, which is nice! But if I understand correctly, it's the same bacterial polishing model from Sep 2024, not a new model.
github.com/nanoporetech...
It supports using --bacteria when polishing an assembly with v5.2.0 data, which is nice! But if I understand correctly, it's the same bacterial polishing model from Sep 2024, not a new model.
myloasm-docs.github.io
myloasm-docs.github.io
myloasm-docs.github.io
In it, I benchmark the new version of Dorado from @nanoporetech.com, which comes with new DNA basecalling models. Short version: big accuracy gains for hac, small improvements for sup.
Check it out for the full results:
rrwick.github.io/2025/05/27/d...
In it, I benchmark the new version of Dorado from @nanoporetech.com, which comes with new DNA basecalling models. Short version: big accuracy gains for hac, small improvements for sup.
Check it out for the full results:
rrwick.github.io/2025/05/27/d...
github.com/nanoporetech...
And for the first time in a year, this includes new DNA basecalling models!
I'll try out these new models (v5.2.0) and compare them to the previous ones (v5.0.0) in a blog post in the near future.

github.com/nanoporetech...
And for the first time in a year, this includes new DNA basecalling models!
I'll try out these new models (v5.2.0) and compare them to the previous ones (v5.0.0) in a blog post in the near future.
www.biorxiv.org/content/10.1...
(1/6)

www.biorxiv.org/content/10.1...
(1/6)
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
If so, you might this little project: github.com/rrwick/conda...
If so, you might this little project: github.com/rrwick/conda...
We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)

We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)
rrwick.github.io/2024/08/16/s...
rrwick.github.io/2024/08/16/s...
rrwick.github.io/2025/02/19/f...
rrwick.github.io/2025/02/19/f...

Ever seen a genome assembly in FASTQ format instead of FASTA? Dorado polish from @nanoporetech.com can do it, and I took a closer look:
rrwick.github.io/2025/02/19/f...
Ever seen a genome assembly in FASTQ format instead of FASTA? Dorado polish from @nanoporetech.com can do it, and I took a closer look:
rrwick.github.io/2025/02/19/f...
rrwick.github.io/2025/02/07/d...
rrwick.github.io/2025/02/07/d...
www.microbiologyresearch.org/content/jour...
(1/4)

www.microbiologyresearch.org/content/jour...
(1/4)
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
github.com/marbl/canu/r...
The notes say this will be the very last release. Many thanks to its devs Brian Walenz and @sergek.bsky.social. Canu may be a bit old and it's certainly not fast, but it makes good assemblies and so I still use it!

github.com/marbl/canu/r...
The notes say this will be the very last release. Many thanks to its devs Brian Walenz and @sergek.bsky.social. Canu may be a bit old and it's certainly not fast, but it makes good assemblies and so I still use it!
🚀 Dorado polish command (experimental) for improving draft assemblies – faster & more accurate than Medaka
⚡ Faster modified base calling models
🔧 Usability & accuracy improvements: PolyA, Barcoding, 6mA calling
github.com/nanoporetech...



