
Past: BS-MS at IISER Pune, India. MS thesis at CES, IISc.
Webpage: https://thepandalorian.github.io
he/him
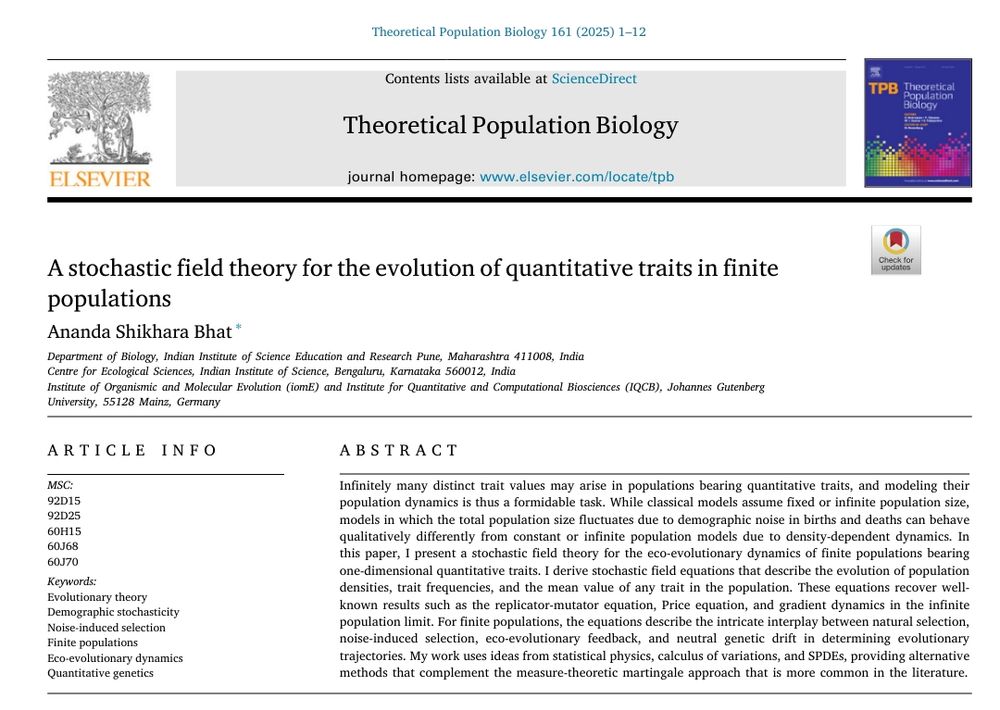
www.sciencedirect.com/science/arti...
I'm super proud of this y'all :')
#scisky 🧪 🧬 #mathsky #physics
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
Title: The Evolution of Dependence and Cohesion in Incipient Endosymbioses.
When: 🌟Thursday, 4th December, at 3 PM (Stockholm time zone).
Zoom link: umu.zoom.us/j/68775661118
Webinar schedule: endosymbiosiswebinar.github.io
#Endosymbiosis

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
doi.org/10.1093/jeb/...
#OpenAccess #Eusociality #GroupAdaptation #FormalDarwinism

doi.org/10.1093/jeb/...
#OpenAccess #Eusociality #GroupAdaptation #FormalDarwinism

are you dumb, nature is green ?
are you dumb, nature is green ?
evol.mcmaster.ca/brian/evoldi...
evol.mcmaster.ca/brian/evoldi...
Please share widely!
#QueerInSTEM #DisabledInSTEM #BlackInSTEM #WomenInSTEM #CarersInSTEM #AcademicSky #HigherED
Please share widely!
#QueerInSTEM #DisabledInSTEM #BlackInSTEM #WomenInSTEM #CarersInSTEM #AcademicSky #HigherED
[[email protected]](mailto:[email protected])
Submit a paper to our special issue in @jevbio.bsky.social!
[[email protected]](mailto:[email protected])
Submit a paper to our special issue in @jevbio.bsky.social!


⭐Microbial cross-feeding: coexistence and collapse, spatial patterns and population cycles⭐
Free and open to all:
Zoom link: iite.info/seminar/
Global Times: www.timeanddate.com/worldclock/f...

www.chemistryworld.com/opinion/the-...

www.chemistryworld.com/opinion/the-...

www.chemistryworld.com/opinion/the-...
We encourage young scientists working at the interface between biology and physics to join us at the IESC in Corsica. Website here: intcha26.sciencesconf.org

We encourage young scientists working at the interface between biology and physics to join us at the IESC in Corsica. Website here: intcha26.sciencesconf.org
ats.talentadore.com/apply/postdo...
ats.talentadore.com/apply/postdo...
A quick thread (please excuse any hyperbole, misrepresentation, or strawman arguments and point them out for me lol):
A quick thread (please excuse any hyperbole, misrepresentation, or strawman arguments and point them out for me lol):
www.nature.com/articles/d41...
Absolutely yes. And I'm proud that @asn-amnat.bsky.social has led the way on this with requiring code and data editors checking code for about five years now!

www.nature.com/articles/d41...
Absolutely yes. And I'm proud that @asn-amnat.bsky.social has led the way on this with requiring code and data editors checking code for about five years now!
shorturl.at/p70wY

shorturl.at/p70wY
- is efficient to compute
- has bounded error
- has excellent analytic properties
- has very high accuracy for small values, which occur frequently in applications
- is efficient to compute
- has bounded error
- has excellent analytic properties
- has very high accuracy for small values, which occur frequently in applications

