
Keen on oligotrophs 💫
Post-doc @ MBL 🦀
Previous Knauss Fellow in Arctic Ocean Observing ❄️
https://sarahjtucker.com/

With new SAR11 isolate genomes and time-series metagenomes, we reveal coastal and offshore SAR11 ecotypes, identify associated metabolic traits, and pinpoint selective gene sweeps as a likely evolutionary driver to niche partitioning. 🌊🦠
py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)
Here is a blog that explains my motivation for this and how to schedule a meeting:
merenlab.org/2025/11/16/E...

Here is a blog that explains my motivation for this and how to schedule a meeting:
merenlab.org/2025/11/16/E...
We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)

We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)
Team villagers >> team werewolves 💗🐺
Team villagers >> team werewolves 💗🐺
A few years ago, when I asked the NY Times to spell my affiliation as University of Hawaiʻi at Mānoa, they refused because it went against their style guide.

A few years ago, when I asked the NY Times to spell my affiliation as University of Hawaiʻi at Mānoa, they refused because it went against their style guide.
apply.interfolio.com/176402
apply.interfolio.com/176402
With new SAR11 isolate genomes and time-series metagenomes, we reveal coastal and offshore SAR11 ecotypes, identify associated metabolic traits, and pinpoint selective gene sweeps as a likely evolutionary driver to niche partitioning. 🌊🦠

With new SAR11 isolate genomes and time-series metagenomes, we reveal coastal and offshore SAR11 ecotypes, identify associated metabolic traits, and pinpoint selective gene sweeps as a likely evolutionary driver to niche partitioning. 🌊🦠
Details: qevomicrolab.org/Documents/Gr...
Apply by October 1 here: forms.gle/38pPS1Ky84HB...

Details: qevomicrolab.org/Documents/Gr...
Apply by October 1 here: forms.gle/38pPS1Ky84HB...
Here, we assessed the contribution of natural transformation to the acquisition of novel genes. See preprint thread. @molbioevol.bsky.social #microsky #evobio
Here, we assessed the contribution of natural transformation to the acquisition of novel genes. See preprint thread. @molbioevol.bsky.social #microsky #evobio
It is a compound verb formed from two words: verschlimmern ("to make worse") and verbessern ("to improve")
The corresponding noun is die Verschlimmbesserung.

If you’ve recently earned a Ph.D. in any scientific field and want to pursue independent, transdisciplinary research, consider applying.
Deadline: October 1, 2025
Apply here: santafe.edu/sfifellowship

If you’ve recently earned a Ph.D. in any scientific field and want to pursue independent, transdisciplinary research, consider applying.
Deadline: October 1, 2025
Apply here: santafe.edu/sfifellowship
Interested in marine microbial metabolites 🌊🦠? If so, check out this new pub., led by Bryndan Durham & Winn Johnson, that emerged from C-CoMP's Labile DOM workshop: An ecological framework for microbial metabolites in the ocean ecosystem.
tinyurl.com/54bu2xm7

Interested in marine microbial metabolites 🌊🦠? If so, check out this new pub., led by Bryndan Durham & Winn Johnson, that emerged from C-CoMP's Labile DOM workshop: An ecological framework for microbial metabolites in the ocean ecosystem.
tinyurl.com/54bu2xm7
PIs enter your position info here: forms.gle/2XTHBP6CZGEn...
Prospective students (and PIs not recruiting) share the composite list: docs.google.com/spreadsheets...
Please share! 🧪
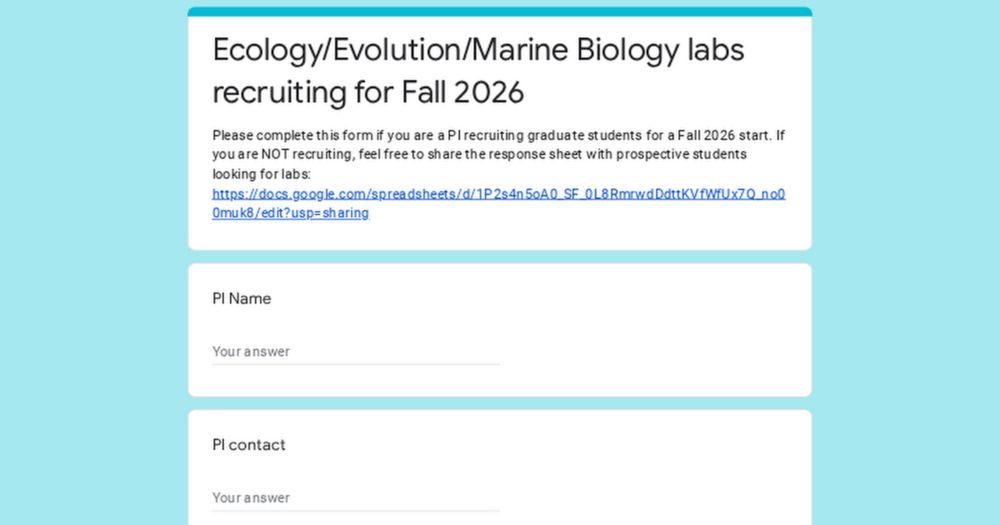
PIs enter your position info here: forms.gle/2XTHBP6CZGEn...
Prospective students (and PIs not recruiting) share the composite list: docs.google.com/spreadsheets...
Please share! 🧪
If you find the resource useful, please also check globdb.org for info on how to cite the underlying resources and tools.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
If you find the resource useful, please also check globdb.org for info on how to cite the underlying resources and tools.
Very important and cool work by @ahoiching.bsky.social @jcamthrash.bsky.social and others
Very important and cool work by @ahoiching.bsky.social @jcamthrash.bsky.social and others
careers.peopleclick.com/careerscp/cl...

careers.peopleclick.com/careerscp/cl...
Culture-supported ecophysiology of the SAR116 clade demonstrates metabolic and spatial niche partitioning academic.oup.com/ismej/advanc... #jcampubs
Culture-supported ecophysiology of the SAR116 clade demonstrates metabolic and spatial niche partitioning academic.oup.com/ismej/advanc... #jcampubs

Application Deadline July 31, 2025

Application Deadline July 31, 2025
We used time-series analyses to identify trends in biogeochemistry and phytoplankton communities across estuarine waters within Kāneʻohe Bay of the Hawaiian island of Oʻahu to the North Pacific Subtropical Gyre. 🏝️

We used time-series analyses to identify trends in biogeochemistry and phytoplankton communities across estuarine waters within Kāneʻohe Bay of the Hawaiian island of Oʻahu to the North Pacific Subtropical Gyre. 🏝️
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵



