Sara Rouhanifard
@srouhanifard.bsky.social
74 followers
91 following
15 posts
Asst. Prof of bioengineering @Northeastern University | RNA modifications | RNA localization | click chemistry | single-molecule studies | www.rouhanifardlab.com
Posts
Media
Videos
Starter Packs
Reposted by Sara Rouhanifard
Reposted by Sara Rouhanifard
Reposted by Sara Rouhanifard
Peng Wu
@pengwu.bsky.social
· Jun 17
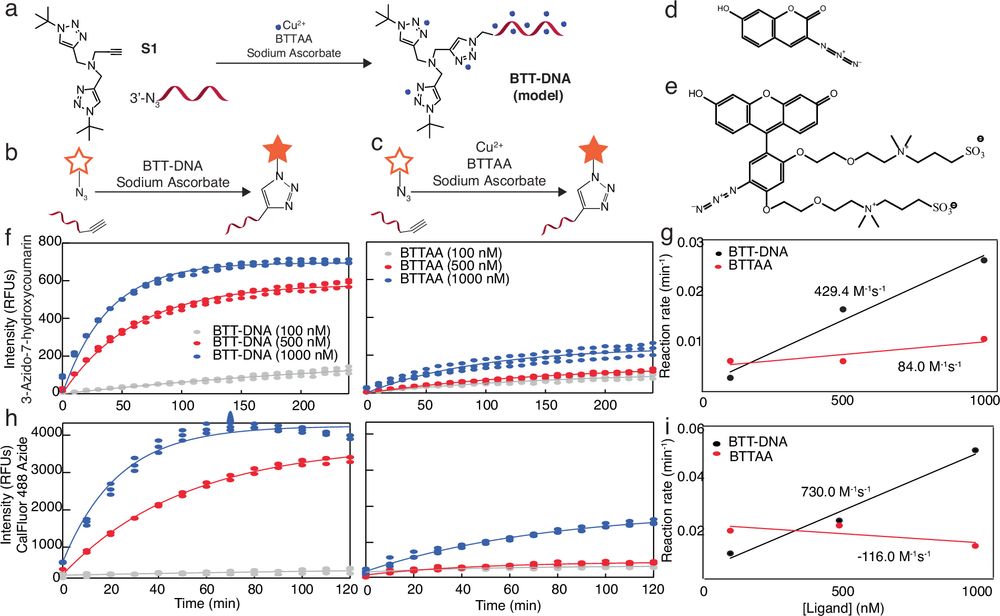
inCu-click: DNA-enhanced ligand enables live-cell, intracellular click chemistry reaction with copper catalyst
Nature Communications - Tracking biomolecules in living cells requires fast but gentle labeling. Here, authors introduce inCu-click, a DNA-conjugated ligand that localizes copper to enable...
rdcu.be
Reposted by Sara Rouhanifard
Arjun Raj
@arjunraj.bsky.social
· Mar 18
Reposted by Sara Rouhanifard
GenomeTDCC
@genometdcc.org
· Mar 10

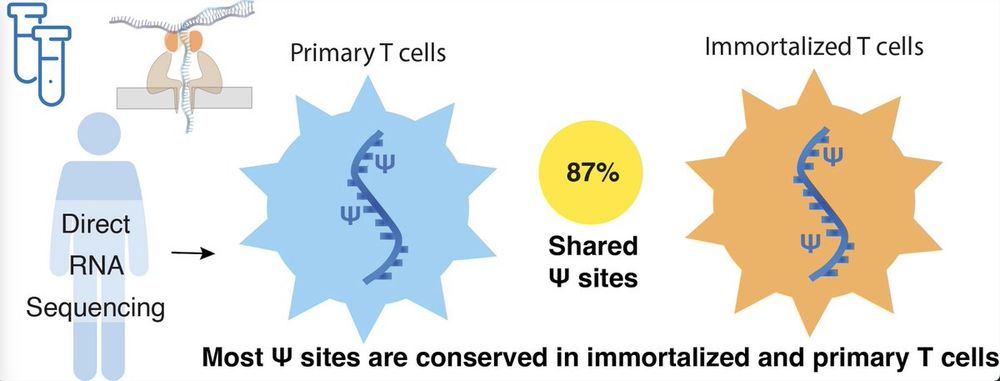
Direct RNA sequencing of primary human T cells reveals the impact of immortalization on mRNA pseudouridine modifications.
Immortalized cell lines are commonly used as proxies for primary cells in human biology research. For example, Jurkat leukemic T cells fundamentally contributed to uncovering T cell signaling, activat...
doi.org
Reposted by Sara Rouhanifard
Reposted by Sara Rouhanifard
Molecular Cell
@cp-molcell.bsky.social
· Feb 19
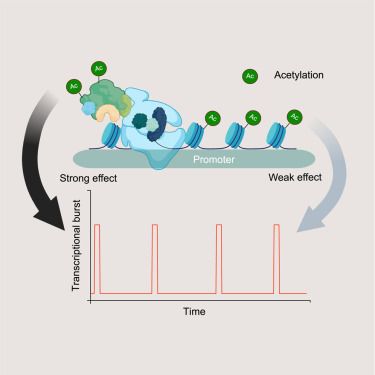
Identification of molecular determinants of gene-specific bursting patterns by high-throughput imaging screens
Sood et al. used high-throughput screening to identify regulators of gene bursting and find both gene- and cell-type-specific effectors. Acetylation alters burst size in a gene-specific manner but is independent of promoter acetylation. Rather,…
dlvr.it
Reposted by Sara Rouhanifard