Aafke Gros
@aafkegros.bsky.social
750 followers
280 following
29 posts
Check out Microscopy Nodes for handling microscopy data in Blender! Now loading .tif and OME-Zarr with big volume support :)
she/her | IPA: ˈafkə ɡrɔs
Posts
Media
Videos
Starter Packs
Reposted by Aafke Gros
Reposted by Aafke Gros
Aafke Gros
@aafkegros.bsky.social
· Aug 27
Reposted by Aafke Gros
Susan Cox
@micoxscopy.bsky.social
· Aug 12

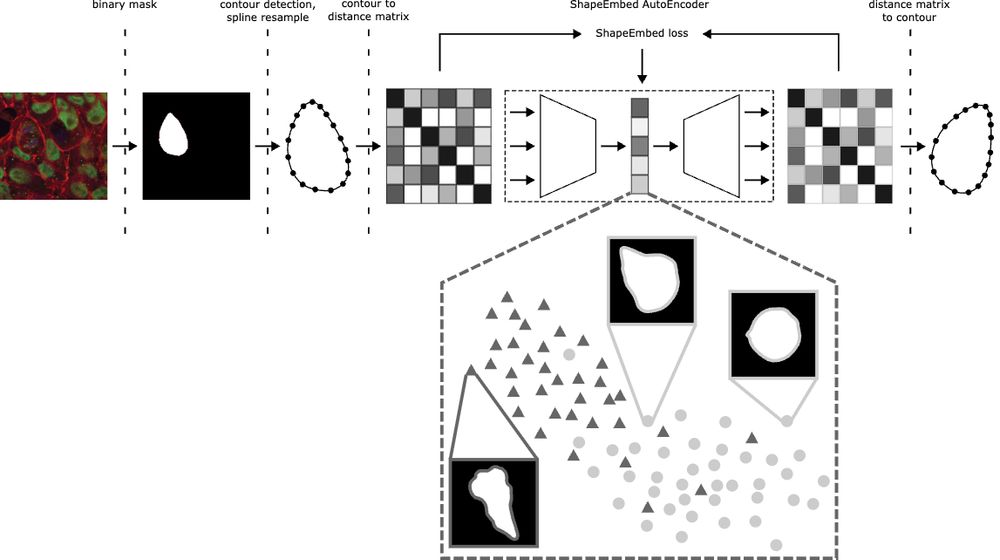
Extracting biological structure and heterogeneity from the nano to the macro scale
Fluorescence microscopy is an essential tool in biology. It has revealed great variability at multiple scales, in macromolecular complexes, cells, and organisms. Understanding this variability will re...
www.biorxiv.org
Reposted by Aafke Gros
Reposted by Aafke Gros
Reposted by Aafke Gros
Reposted by Aafke Gros
Reposted by Aafke Gros
Reposted by Aafke Gros
Reposted by Aafke Gros