Posts
Media
Videos
Starter Packs
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Anaïs Baudot
@anaisbaudot.bsky.social
· Jul 26
Anaïs Baudot
@anaisbaudot.bsky.social
· Jul 26
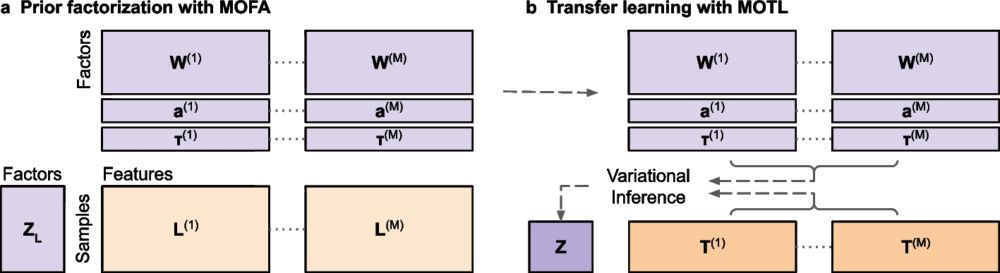
MOTL: enhancing multi-omics matrix factorization with transfer learning - Genome Biology
Joint matrix factorization is popular for extracting lower dimensional representations of multi-omics data but loses effectiveness with limited samples. Addressing this limitation, we introduce MOTL (...
link.springer.com
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot
Reposted by Anaïs Baudot