Anwai Archit
@anwaiarchit.bsky.social
82 followers
150 following
25 posts
PhD Candidate at @cppape.bsky.social lab.
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Anwai Archit
Anwai Archit
@anwaiarchit.bsky.social
· Jul 16
Anwai Archit
@anwaiarchit.bsky.social
· Jul 16
Anwai Archit
@anwaiarchit.bsky.social
· Jul 16
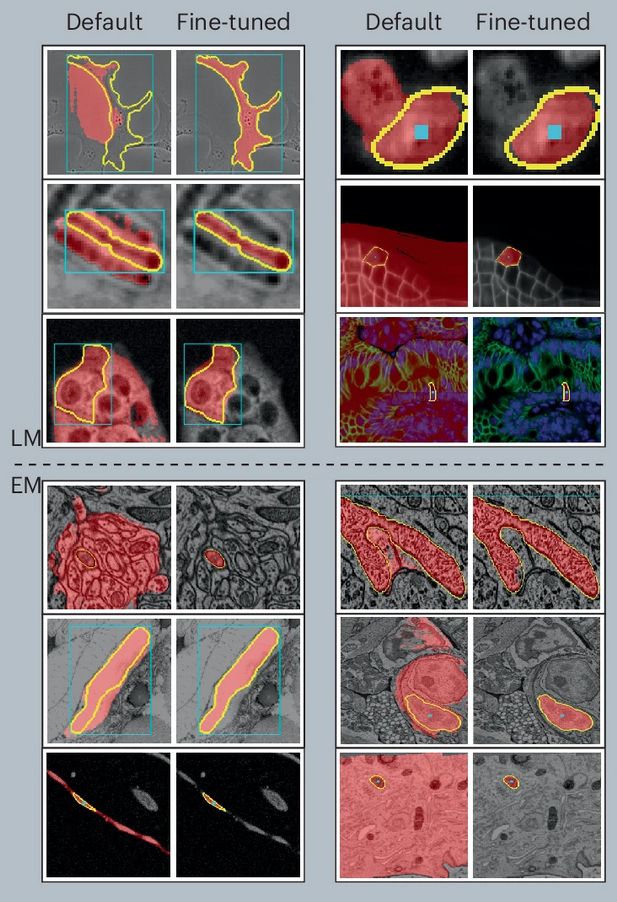
Parameter Efficient Fine-Tuning of Segment Anything Model for Biomedical Imaging
Segmentation is an important analysis task for biomedical images, enabling the study of individual organelles, cells or organs. Deep learning has massively improved segmentation methods, but challenge...
doi.org
Reposted by Anwai Archit
Constantin Pape
@cppape.bsky.social
· Jul 7
Reposted by Anwai Archit
Anwai Archit
@anwaiarchit.bsky.social
· Jun 8
Reposted by Anwai Archit
Nature Methods
@natmethods.nature.com
· Jun 6
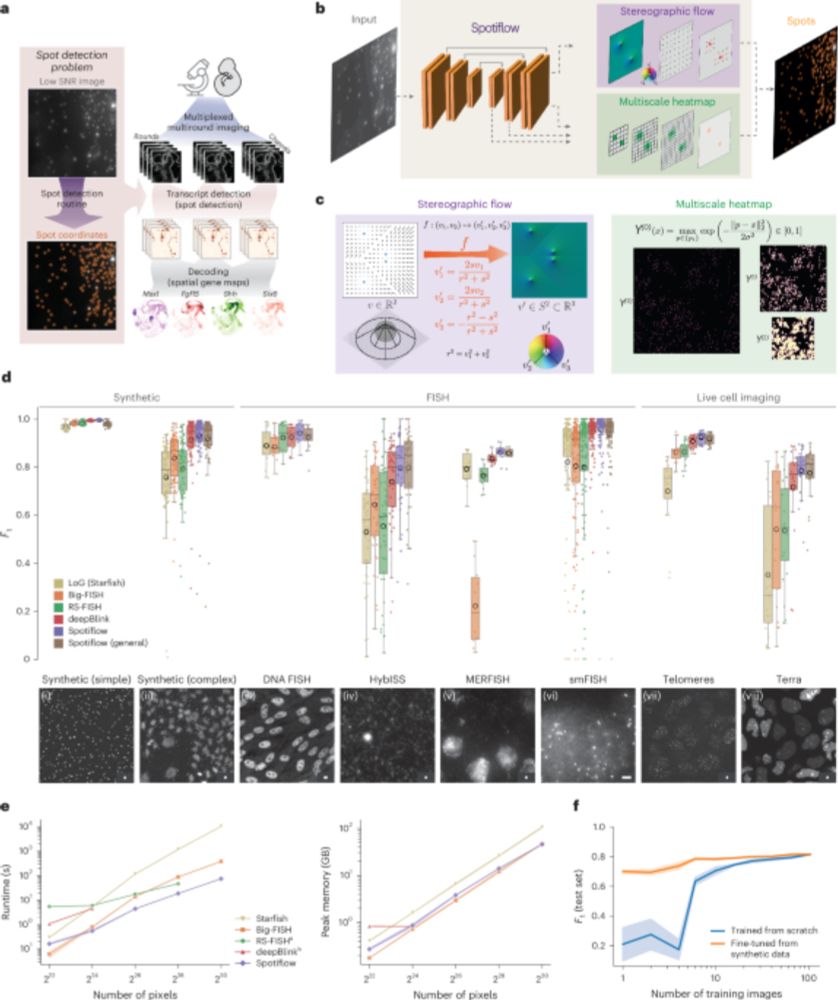
Spotiflow: accurate and efficient spot detection for fluorescence microscopy with deep stereographic flow regression - Nature Methods
Spotiflow uses deep learning for subpixel-accurate spot detection in diverse 2D and 3D images. The improved accuracy offered by Spotiflow enables improved biological insights in both iST and live imag...
www.nature.com
Reposted by Anwai Archit
Constantin Pape
@cppape.bsky.social
· Mar 28
Reposted by Anwai Archit
Reposted by Anwai Archit
Anwai Archit
@anwaiarchit.bsky.social
· Mar 13
Anwai Archit
@anwaiarchit.bsky.social
· Mar 3
Reposted by Anwai Archit
Reposted by Anwai Archit
Reposted by Anwai Archit
Reposted by Anwai Archit
Reposted by Anwai Archit