Ahmet Sarigun
@asarigun.bsky.social
27 followers
110 following
8 posts
curious explorer - https://asarigun.github.io/
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Ahmet Sarigun
Matthieu da Costa
@mdc-biocat.bsky.social
· Aug 31
Reposted by Ahmet Sarigun
Reposted by Ahmet Sarigun
Reposted by Ahmet Sarigun
Ahmet Sarigun
@asarigun.bsky.social
· Jun 26

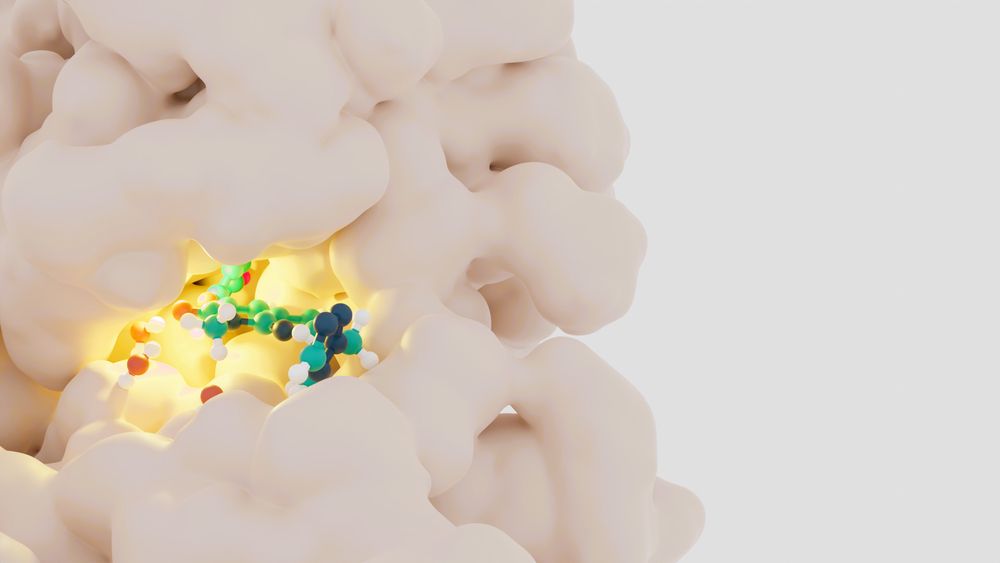
PocketVina Enables Scalable and Highly Accurate Physically Valid Docking through Multi-Pocket Conditioning
Sampling physically valid ligand-binding poses remains a major challenge in molecular docking, particularly for unseen or structurally diverse targets. We introduce PocketVina, a fast and memory-effic...
arxiv.org
Reposted by Ahmet Sarigun
Ahmet Sarigun
@asarigun.bsky.social
· Feb 3
Reposted by Ahmet Sarigun
Reposted by Ahmet Sarigun
Pia Rautenstrauch
@prauten.bsky.social
· Jan 24

Metrics Matter: Why We Need to Stop Using Silhouette in Single-Cell Benchmarking
Current-day single-cell studies comprise complex data sets affected by nested batch effects caused by technical and biological factors, relying on advanced integration methods. Silhouette is an establ...
www.biorxiv.org