Pia Rautenstrauch
@prauten.bsky.social
110 followers
190 following
11 posts
Computer Science PhD Student at @humboldtuni.bsky.social and @mdc-berlin.bsky.social | Data Science | Machine learning | AI | Bioinformatics | Genomics | Single-Cell Biology
Posts
Media
Videos
Starter Packs
Pinned
Pia Rautenstrauch
@prauten.bsky.social
· Jan 24

Metrics Matter: Why We Need to Stop Using Silhouette in Single-Cell Benchmarking
Current-day single-cell studies comprise complex data sets affected by nested batch effects caused by technical and biological factors, relying on advanced integration methods. Silhouette is an establ...
www.biorxiv.org
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Tuomo Hartonen
@tuomohartonen.bsky.social
· Aug 27

Cross-biobank generalizability and accuracy of electronic health record-based predictors compared to polygenic scores - Nature Genetics
Comparison of electronic health record-based phenotype risk scores (PheRS) and polygenic scores (PGS) across 13 common diseases and three biobank-based studies indicates that PheRS and PGS may provide...
www.nature.com
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Pia Rautenstrauch
@prauten.bsky.social
· Aug 12
Pia Rautenstrauch
@prauten.bsky.social
· Aug 12
Pia Rautenstrauch
@prauten.bsky.social
· Aug 12
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Liana Lareau
@lianafaye.bsky.social
· Aug 7

Protein language models reveal evolutionary constraints on synonymous codon choice
Evolution has shaped the genetic code, with subtle pressures leading to preferences for some synonymous codons over others. Codons are translated at different speeds by the ribosome, imposing constrai...
www.biorxiv.org
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
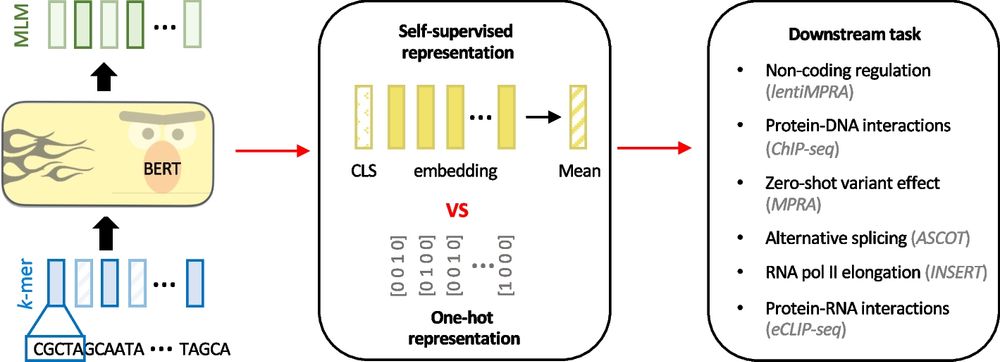
Evaluating the representational power of pre-trained DNA language models for regulatory genomics - Genome Biology
Background The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis-regulatory patterns in the non-coding genome without requiring labels of ...
genomebiology.biomedcentral.com
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch
Reposted by Pia Rautenstrauch