Emma Dann
@emmamarydann.bsky.social
760 followers
470 following
3 posts
Postdoc fellow @ Stanford & Gladstone Institutes
Core team @scverse-team.bsky.social
Bringing the single-cell genomics in human complex trait genetics
https://emdann.github.io/
Posts
Media
Videos
Starter Packs
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
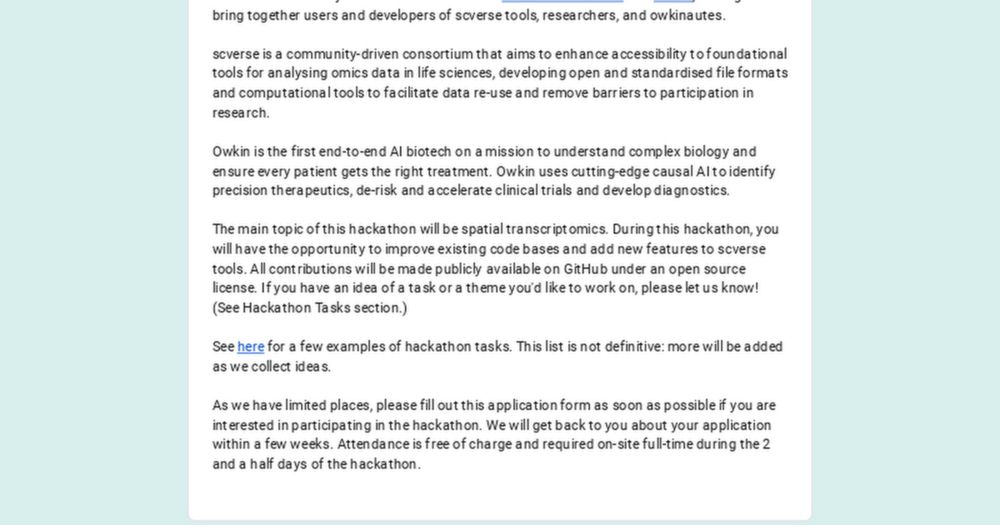
scverse
@scverse.bsky.social
· Aug 2
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Jonathan Pritchard
@jkpritch.bsky.social
· Jan 27
Jonathan Pritchard
@jkpritch.bsky.social
· Jan 26

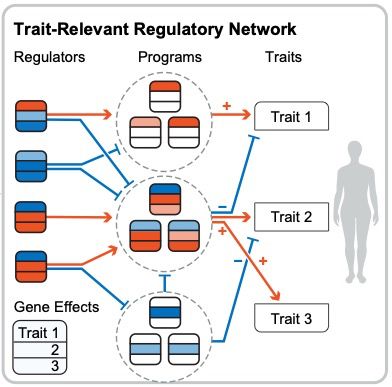
Causal modeling of gene effects from regulators to programs to traits: integration of genetic associations and Perturb-seq
Genetic association studies provide a unique tool for identifying causal links from genes to human traits and diseases. However, it is challenging to determine the biological mechanisms underlying mos...
www.biorxiv.org
Reposted by Emma Dann
Jonathan Pritchard
@jkpritch.bsky.social
· Jan 24
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann
Jeff Spence
@jeffspence.github.io
· Dec 17

Specificity, length, and luck: How genes are prioritized by rare and common variant association studies
Standard genome-wide association studies (GWAS) and rare variant burden tests are essential tools for identifying trait-relevant genes. Although these methods are conceptually similar, we show by anal...
www.biorxiv.org
Reposted by Emma Dann
Reposted by Emma Dann
Reposted by Emma Dann