Constantin Ahlmann-Eltze
@const-ae.bsky.social
2K followers
580 following
58 posts
Postdoc at UCL with James Reading. Previously at EMBL working with Wolfgang Huber. Biostats, R, cancer immunology
Posts
Media
Videos
Starter Packs
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Carl Zimmer
@carlzimmer.com
· Aug 4
Reposted by Constantin Ahlmann-Eltze
Nature Methods
@natmethods.nature.com
· Aug 4
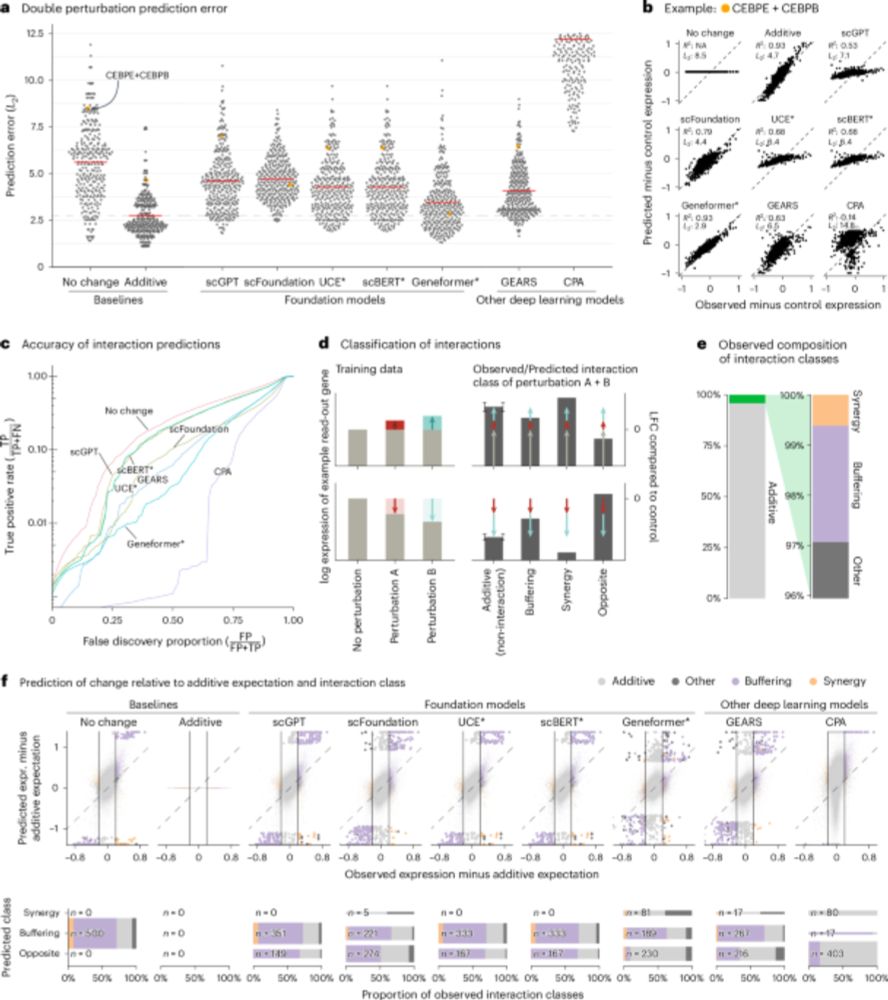
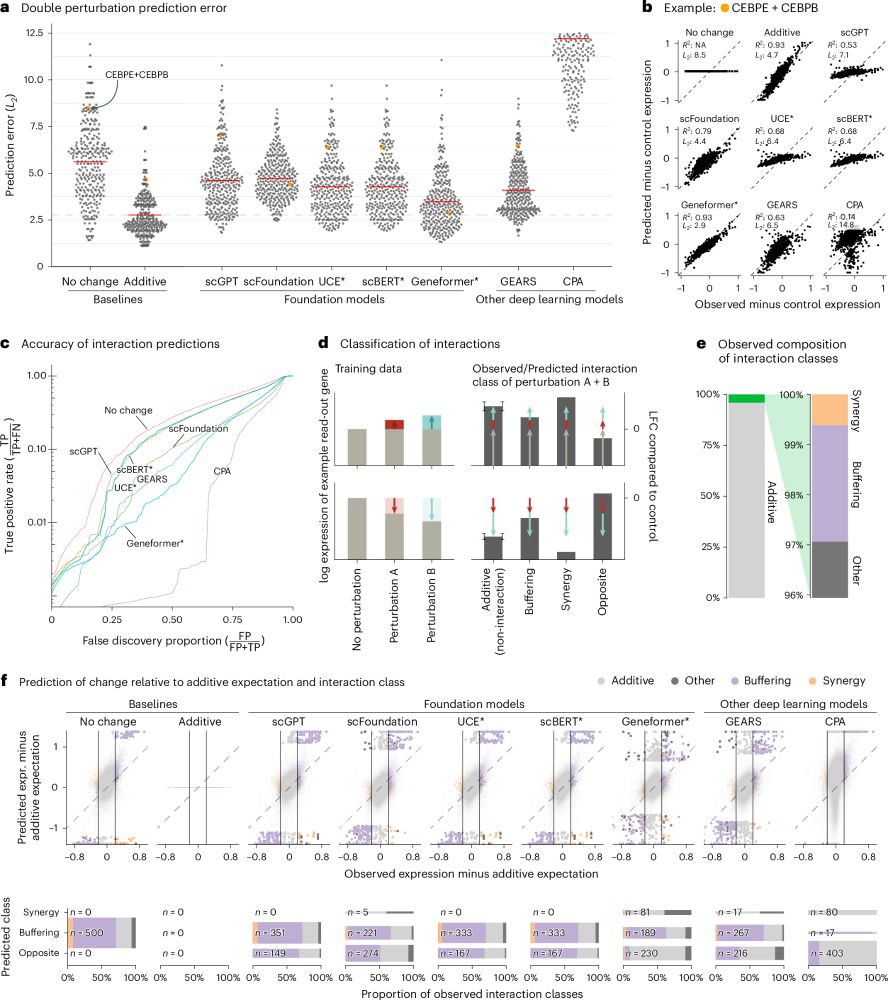
Deep-learning-based gene perturbation effect prediction does not yet outperform simple linear baselines - Nature Methods
The analysis presented in this Brief Communication shows that, despite their complexity, current deep learning models do not outperform linear baselines in predicting gene perturbation effects, thus e...
www.nature.com
Reposted by Constantin Ahlmann-Eltze
Eric Kernfeld
@ekernf01.bsky.social
· Jul 27
A recap of virtual cell releases circa June 2025
In October 2024, I twote that “something is deeply wrong” with what we now call virtual cell models. A lot has happened since then: modelers are advancing new architectures and mining new sources of i...
ekernf01.github.io
Reposted by Constantin Ahlmann-Eltze
Helena Lucia Crowell
@helucro.bsky.social
· Jun 27

Tracing colorectal malignancy transformation from cell to tissue scale
The transformation of normal intestinal epithelium into colorectal cancer (CRC) involves coordinated changes across molecular, cellular, and architectural scales; yet, how these layers integrate remai...
www.biorxiv.org
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Reposted by Constantin Ahlmann-Eltze
Lorenz Adlung Lab
@adlunglab.com
· Mar 19
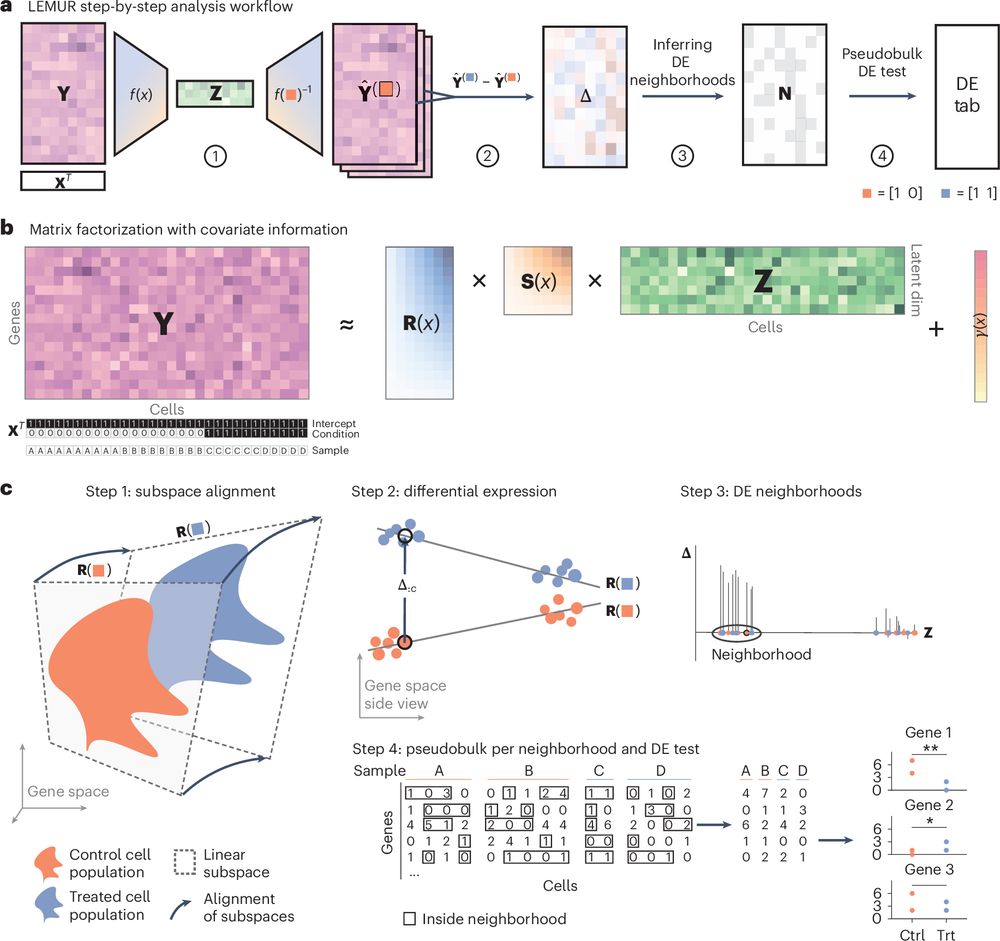
Analysis of multi-condition single-cell data with latent embedding multivariate regression
Nature Genetics - Latent embedding multivariate regression models multi-condition single-cell RNA-seq using a continuous latent space, enabling data integration, per-cell gene expression prediction...
doi.org