Stein Aerts
@steinaerts.bsky.social
2.8K followers
640 following
33 posts
Computational biologist interested in deciphering the genomic regulatory code at vib.ai
Posts
Media
Videos
Starter Packs
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
Reposted by Stein Aerts
David Kelley
@drkbio.bsky.social
· Jul 21

Borzoi-informed fine mapping improves causal variant prioritization in complex trait GWAS
Genome-wide association studies (GWAS) have identified thousands of trait-associated loci. Prioritizing causal variants within these loci is critical for characterizing trait biology. Statistical fine...
www.biorxiv.org
Reposted by Stein Aerts
Carl de Boer
@carldeboer.bsky.social
· Jul 11
Reposted by Stein Aerts
Anamaria Elek
@aelek.bsky.social
· Jul 6
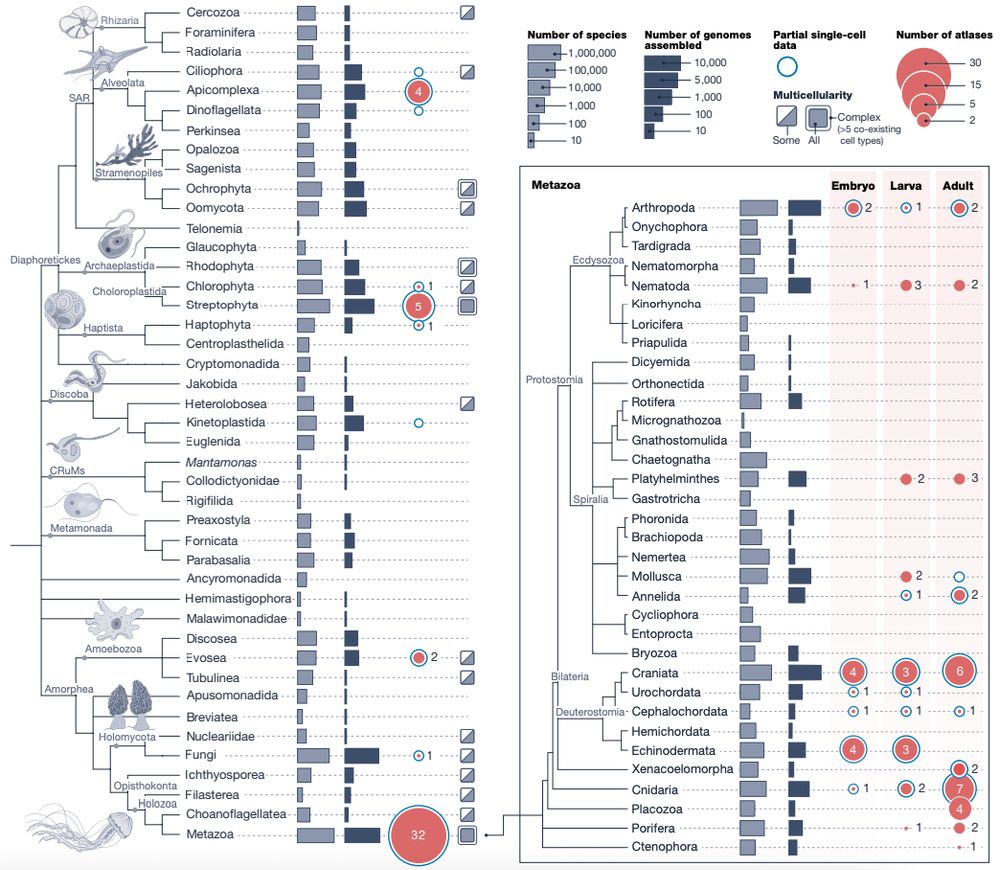
Decoding cnidarian cell type gene regulation
Animal cell types are defined by differential access to genomic information, a process orchestrated by the combinatorial activity of transcription factors that bind to cis -regulatory elements (CREs) to control gene expression. However, the regulatory logic and specific gene networks that define cell identities remain poorly resolved across the animal tree of life. As early-branching metazoans, cnidarians can offer insights into the early evolution of cell type-specific genome regulation. Here, we profiled chromatin accessibility in 60,000 cells from whole adults and gastrula-stage embryos of the sea anemone Nematostella vectensis. We identified 112,728 CREs and quantified their activity across cell types, revealing pervasive combinatorial enhancer usage and distinct promoter architectures. To decode the underlying regulatory grammar, we trained sequence-based models predicting CRE accessibility and used these models to infer ontogenetic relationships among cell types. By integrating sequence motifs, transcription factor expression, and CRE accessibility, we systematically reconstructed the gene regulatory networks that define cnidarian cell types. Our results reveal the regulatory complexity underlying cell differentiation in a morphologically simple animal and highlight conserved principles in animal gene regulation. This work provides a foundation for comparative regulatory genomics to understand the evolutionary emergence of animal cell type diversity. ### Competing Interest Statement The authors have declared no competing interest. European Research Council, https://ror.org/0472cxd90, ERC-StG 851647 Ministerio de Ciencia e Innovación, https://ror.org/05r0vyz12, PID2021-124757NB-I00, FPI Severo Ochoa PhD fellowship European Union, https://ror.org/019w4f821, Marie Skłodowska-Curie INTREPiD co-fund agreement 75442, Marie Skłodowska-Curie grant agreement 101031767
www.biorxiv.org
Reposted by Stein Aerts
Oliver Stegle
@oliverstegle.bsky.social
· Jun 26

Postdoctoral Researcher in Computational Genetics
The research group of Oliver Stegle looks for a postdoctoral researcher to join a collaborative project with GSK with the goal to apply computational methods to investigate the effects of rare variant...
embl.wd103.myworkdayjobs.com
Reposted by Stein Aerts
Žiga Avsec
@avsecz.bsky.social
· Jun 25
Reposted by Stein Aerts
Reposted by Stein Aerts