Alexandra P
@alexanrna.bsky.social
140 followers
410 following
12 posts
🇸🇰 PhD student @ KU Leuven and VIB 🇧🇪
Genetics, Bioinformatics, and everything in between (she/her)
Posts
Media
Videos
Starter Packs
Reposted by Alexandra P
Reposted by Alexandra P
Reposted by Alexandra P
scverse
@scverse.bsky.social
· 28d
Reposted by Alexandra P
Reposted by Alexandra P
Reposted by Alexandra P
FlyBase
@flybase.bsky.social
· Jun 3
Reposted by Alexandra P
Reposted by Alexandra P
Reposted by Alexandra P
Reposted by Alexandra P
Reposted by Alexandra P
Reposted by Alexandra P
Niklas Kempynck
@niklaskemp.bsky.social
· Feb 14
VIB.AI
@vibai.bsky.social
· Feb 14
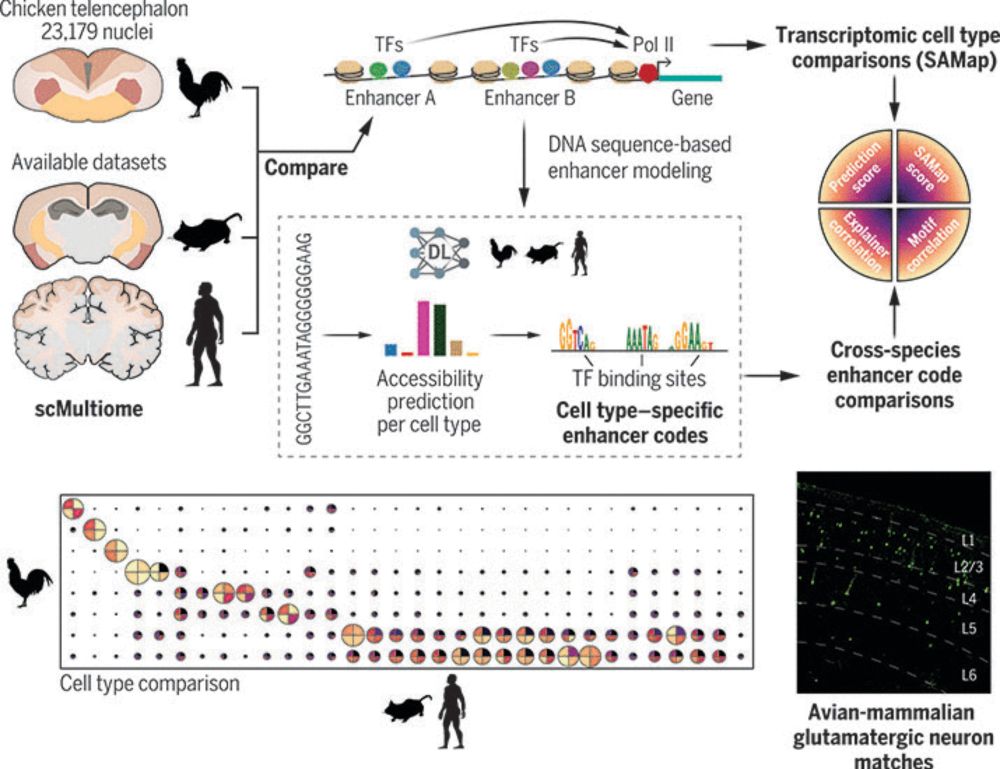
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Combinations of transcription factors govern the identity of cell types, which is reflected by genomic enhancer codes. We used deep learning to characterize these enhancer codes and devised three metr...
www.science.org