Seppe De Winter
@seppedewinter.bsky.social
64 followers
110 following
5 posts
PhD researcher at aertslab VIB-AI KU Leuven.
Posts
Media
Videos
Starter Packs
Reposted by Seppe De Winter
Reposted by Seppe De Winter
Niklas Kempynck
@niklaskemp.bsky.social
· May 21
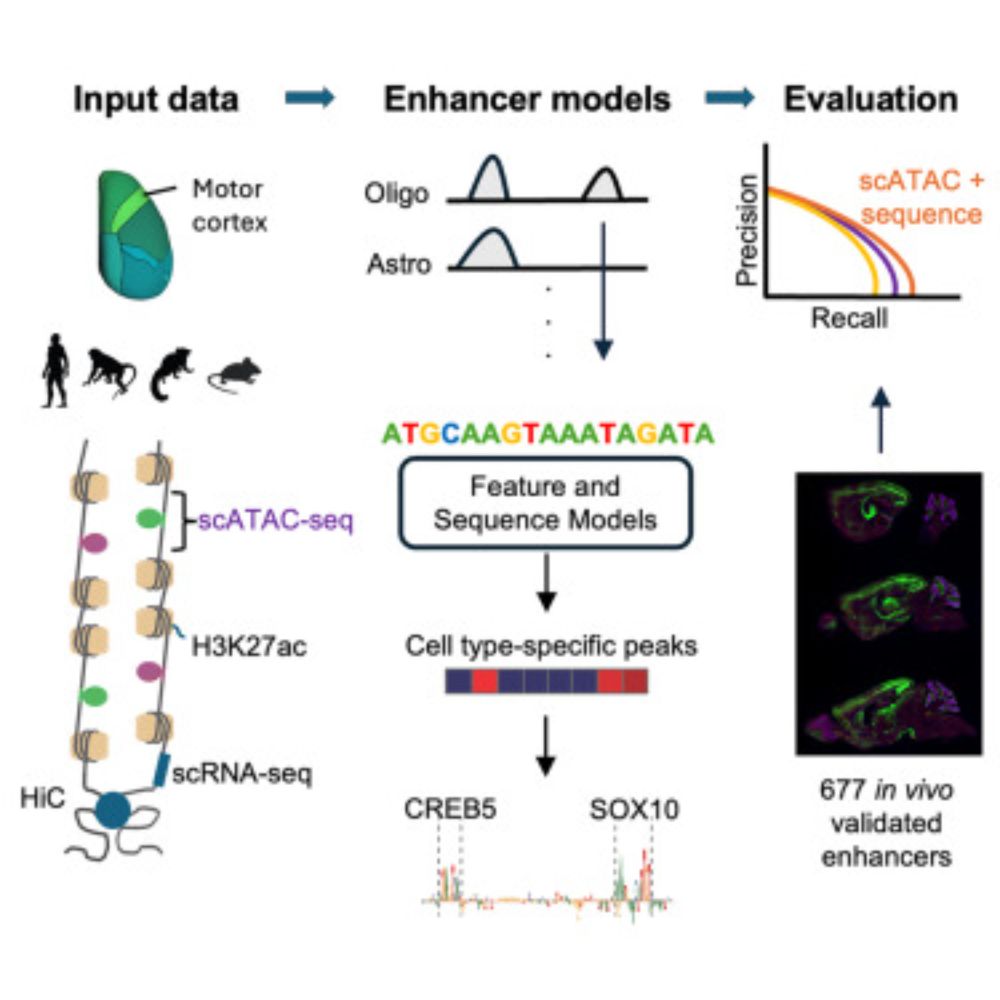
Evaluating methods for the prediction of cell-type-specific enhancers in the mammalian cortex
Johansen et al. report the results of a community challenge to predict functional
enhancers targeting specific brain cell types. By comparing multi-omics machine learning
approaches using in vivo data...
www.cell.com
Reposted by Seppe De Winter
Stein Aerts
@steinaerts.bsky.social
· May 21
Reposted by Seppe De Winter
Stein Aerts
@steinaerts.bsky.social
· Apr 4

CREsted: modeling genomic and synthetic cell type-specific enhancers across tissues and species
Sequence-based deep learning models have become the state of the art for the analysis of the genomic regulatory code. Particularly for transcriptional enhancers, deep learning models excel at decipher...
www.biorxiv.org
Reposted by Seppe De Winter
Reposted by Seppe De Winter
Reposted by Seppe De Winter
Reposted by Seppe De Winter
Reposted by Seppe De Winter
Reposted by Seppe De Winter
Reposted by Seppe De Winter
VIB.AI
@vibai.bsky.social
· Feb 14
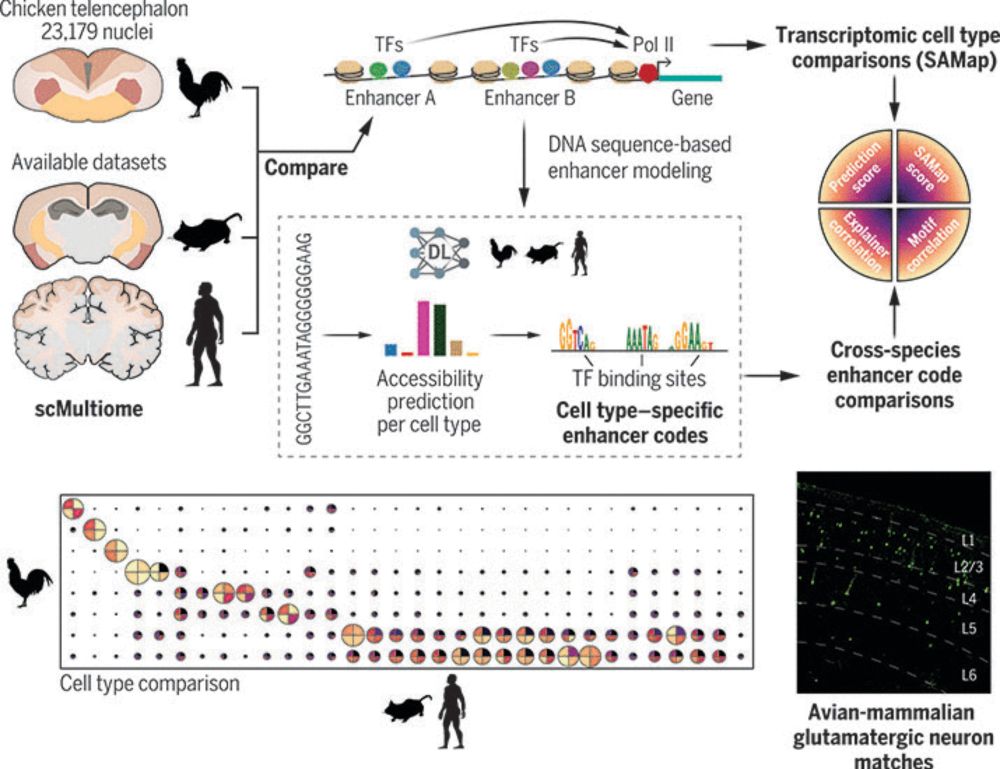
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Combinations of transcription factors govern the identity of cell types, which is reflected by genomic enhancer codes. We used deep learning to characterize these enhancer codes and devised three metr...
www.science.org
Reposted by Seppe De Winter
Stein Aerts
@steinaerts.bsky.social
· Feb 14
Reposted by Seppe De Winter
Niklas Kempynck
@niklaskemp.bsky.social
· Feb 14
VIB.AI
@vibai.bsky.social
· Feb 14

Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Combinations of transcription factors govern the identity of cell types, which is reflected by genomic enhancer codes. We used deep learning to characterize these enhancer codes and devised three metr...
www.science.org