David Bartel's Lab
@bartellab.bsky.social
470 followers
150 following
11 posts
David Bartel's lab @WhiteheadInst @MIT @HHMI | microRNAs, mRNAs, and other RNAs
Posts
Media
Videos
Starter Packs
Reposted by David Bartel's Lab
Eugene Valkov
@eugenevalkov.bsky.social
· Aug 13

Multivalent interactions with CCR4–NOT and PABPC1 determine mRNA repression efficiency by tristetraprolin - Nature Communications
Deadenylation leads to mRNA decay, with PABPC1 protecting the poly(A) tail, while tristetraprolin and CCR4–NOT promote deadenylation. Here, the authors describe how these three proteins interact to re...
www.nature.com
Reposted by David Bartel's Lab
Iain Cheeseman
@iaincheeseman.bsky.social
· Jul 23

Global inhibition of deadenylation stabilizes the transcriptome in mitotic cells
In the presence of cell division errors, mammalian cells can pause in mitosis for tens of hours with little to no transcription, while still requiring continued translation for viability. These unique...
www.biorxiv.org
David Bartel's Lab
@bartellab.bsky.social
· Mar 15
Reposted by David Bartel's Lab
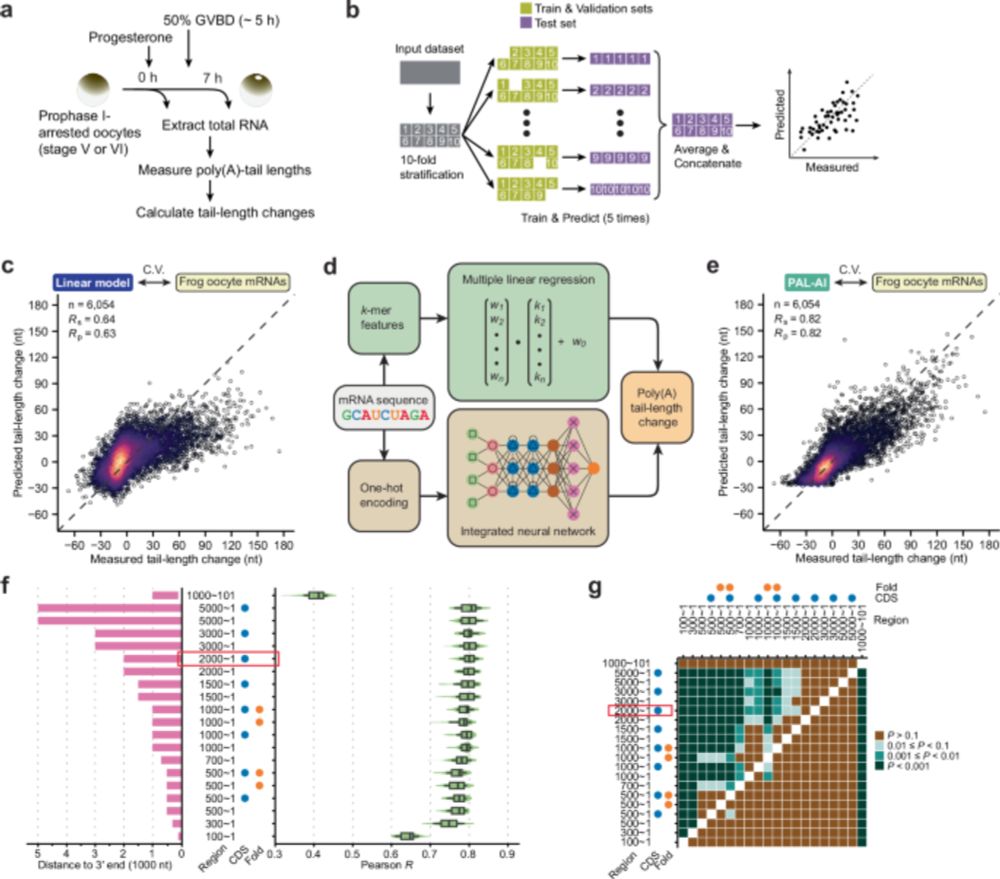
Peter Wang
@wyppeter.bsky.social
· Jan 1
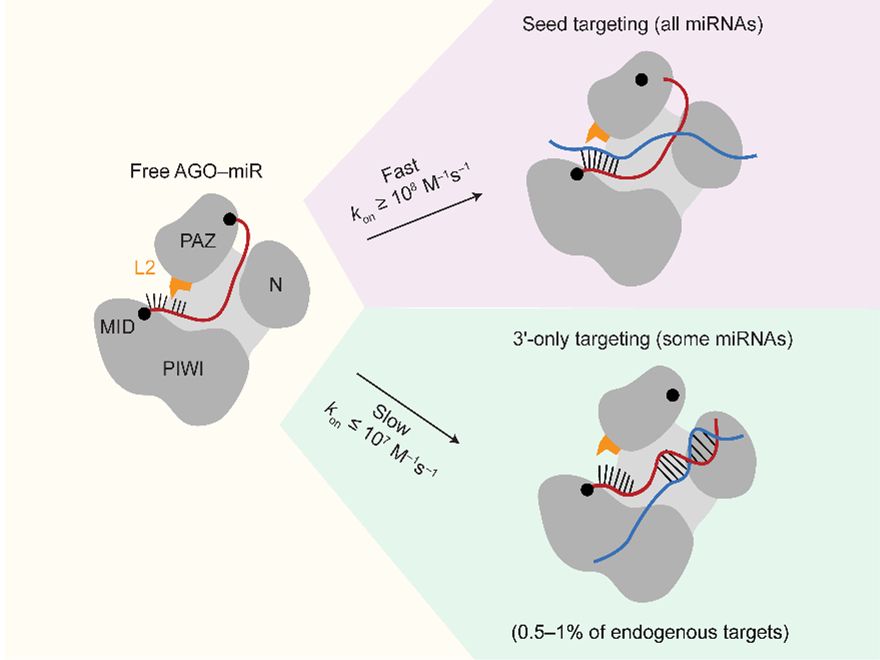
Seychelle Vos
@voslab.org
· Jan 1
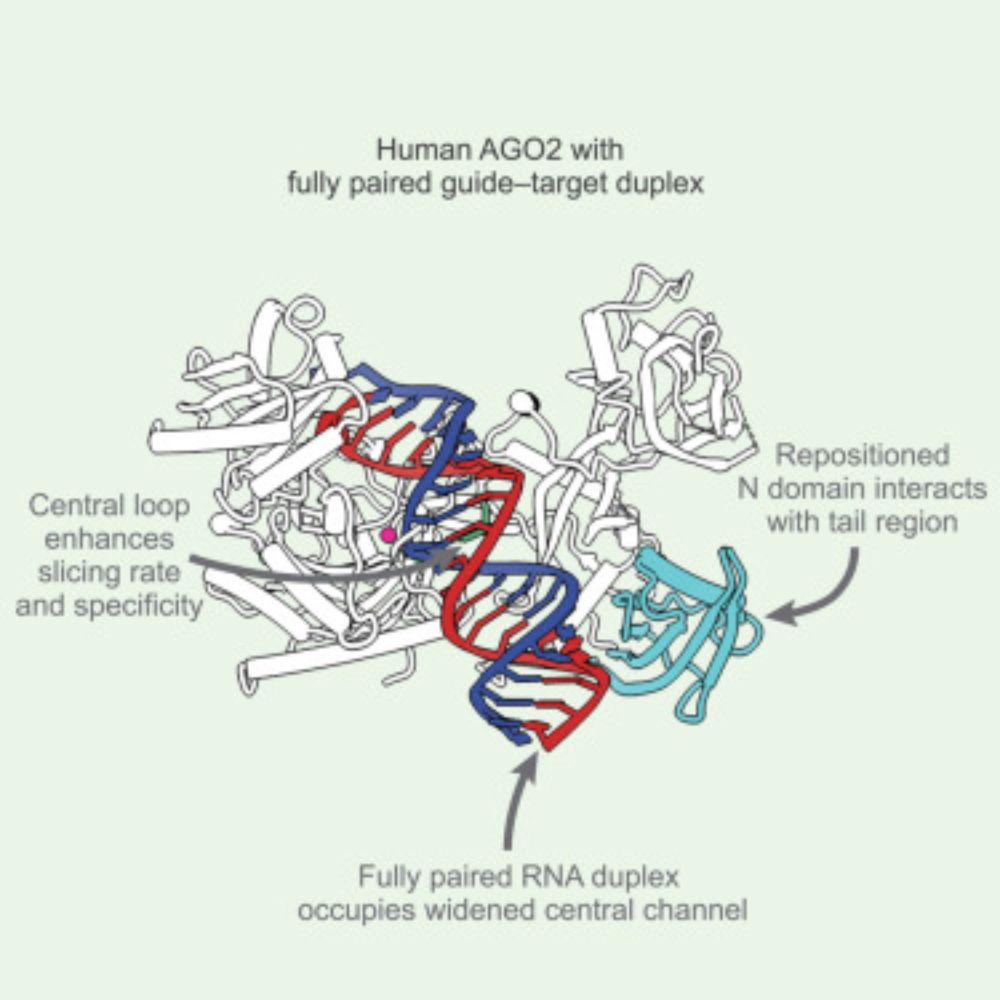
The structural basis for RNA slicing by human Argonaute2
Mohamed et al. report the cryoelectron microscopy structure of human AGO2 with fully paired guide RNA. Their analysis reveals the structural basis for the slicing activity that drives RNAi, showing that the slicing-competent conformation is achieved by domain movements and RNA-protein contacts distinct from those of conformational intermediates and prokaryotic homologs.
tinyurl.com
David Bartel's Lab
@bartellab.bsky.social
· Dec 18
David Bartel's Lab
@bartellab.bsky.social
· Dec 18