Bornberglab
@bornberglab.bsky.social
420 followers
360 following
16 posts
Molecular evolution and bioinformatics at Uni Muenster, Germany. Protein evolution | domains | de novo genes | comparative evolutionary genomics
http://bornberglab.org/
Posts
Media
Videos
Starter Packs
Reposted by Bornberglab
Reposted by Bornberglab
Reposted by Bornberglab
Reposted by Bornberglab
Bornberglab
@bornberglab.bsky.social
· Jul 22
Reposted by Bornberglab
Reposted by Bornberglab
Bornberglab
@bornberglab.bsky.social
· Apr 22
Bornberglab
@bornberglab.bsky.social
· Apr 22

Expression of De Novo Open Reading Frames in Natural Populations of Drosophila melanogaster
We show that newly-evolved, expressed open reading frames (neORFs) identified in a set of inbred Drosophila melanogaster lines are also expressed in multiple tissues and developmental stages of poole...
doi.org
Reposted by Bornberglab
Colin Jackson
@cjjackson.bsky.social
· Apr 15

Fitness Landscape Ruggedness Arises from Biophysical Complexity
Epistasis, in which the effect of a mutation depends on the genetic background it is introduced into, drives protein evolution and design by shaping the fitness landscape. Quantifying how constrained ...
www.biorxiv.org
Reposted by Bornberglab
Reposted by Bornberglab
Bornberglab
@bornberglab.bsky.social
· Apr 3
Reposted by Bornberglab
Reposted by Bornberglab
SPP 2349 Gevol
@gevol.bsky.social
· Apr 2
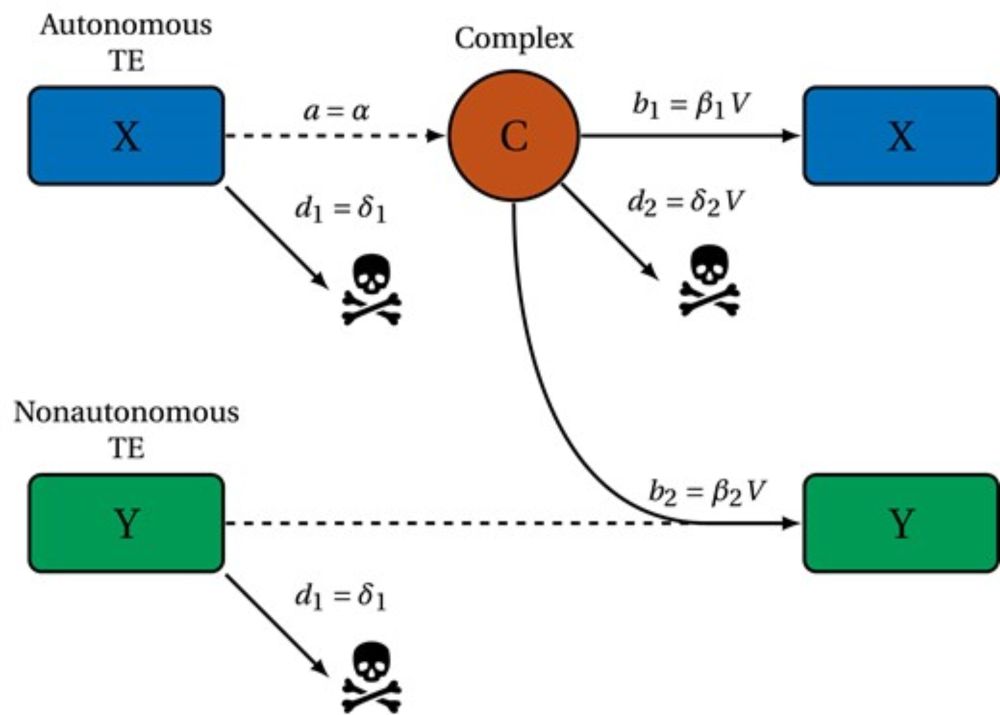
Maintenance of long-term transposable element activity through regulation by nonautonomous elements
Abstract. Transposable elements are DNA sequences that can move and replicate within genomes. Broadly, there are 2 types: autonomous elements, which encode
doi.org