Junhong Choi
@choijunhong.bsky.social
300 followers
310 following
22 posts
PI at MSKCC. Synthetic (Developmental) Biology + Molecular Recording + Genomics Tech Dev. Words like physicality of information make my heart go a little faster.
Posts
Media
Videos
Starter Packs
Reposted by Junhong Choi
Reposted by Junhong Choi
Sternberg Lab
@sternberglab.bsky.social
· May 28
Sternberg Lab
@sternberglab.bsky.social
· Mar 26

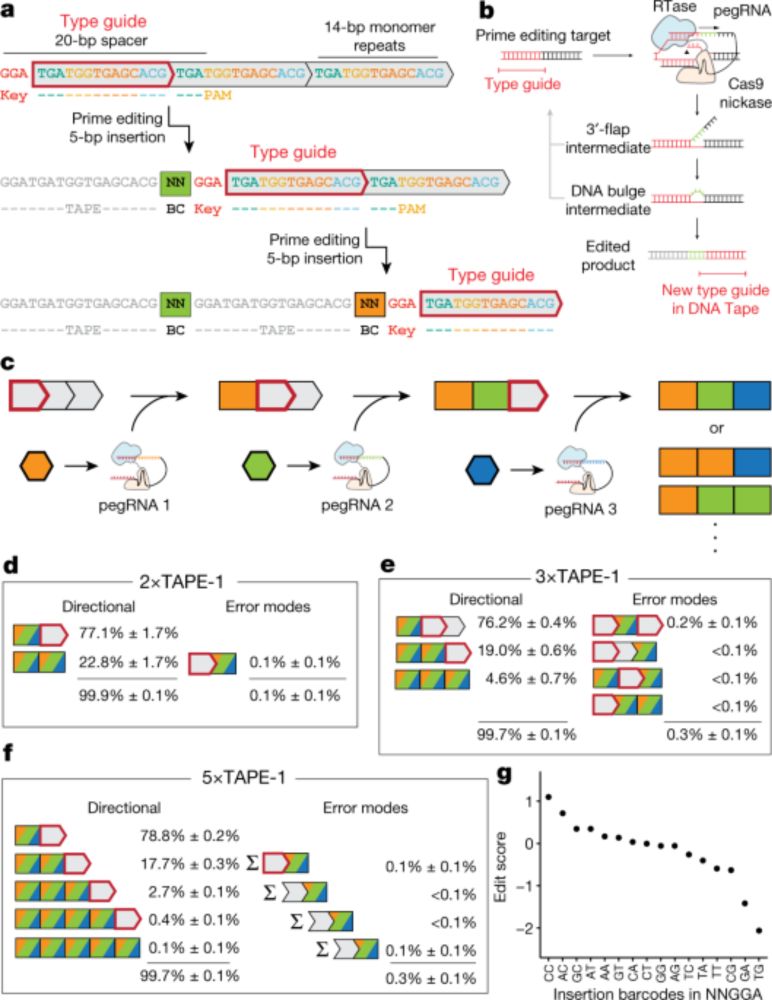
Protein-primed DNA homopolymer synthesis by an antiviral reverse transcriptase
Bacteria defend themselves from viral predation using diverse immune systems, many of which sense and target foreign DNA for degradation. Defense-associated reverse transcriptase (DRT) systems provide...
www.biorxiv.org
Junhong Choi
@choijunhong.bsky.social
· May 28
Junhong Choi
@choijunhong.bsky.social
· May 27
Junhong Choi
@choijunhong.bsky.social
· May 27

Single-cell mapping of lineage and identity in direct reprogramming - Nature
Combinatorial tagging of single cells using expressed DNA barcodes, delivered by a lentiviral vector, is used to track individual cells and reconstruct their lineages and trajectories during cell fate...
doi.org
Junhong Choi
@choijunhong.bsky.social
· May 27

Recording morphogen signals reveals mechanisms underlying gastruloid symmetry breaking - Nature Cell Biology
Toettcher, McNamara and colleagues use synthetic ‘signal-recording’ gene circuits on mouse gastruloids and find that cell sorting rearranges patchy domains of Wnt activity into a single pole, which de...
doi.org
Junhong Choi
@choijunhong.bsky.social
· May 27
Silvia Domcke
@sdomcke.bsky.social
· May 25

Barcoded monoclonal embryoids are a potential solution to confounding bottlenecks in mosaic organoid screens
Genetic screens in organoids hold tremendous promise for accelerating discoveries at the intersection of genomics and developmental biology. Embryoid bodies (EBs) are self-organizing multicellular str...
www.biorxiv.org
Junhong Choi
@choijunhong.bsky.social
· May 27

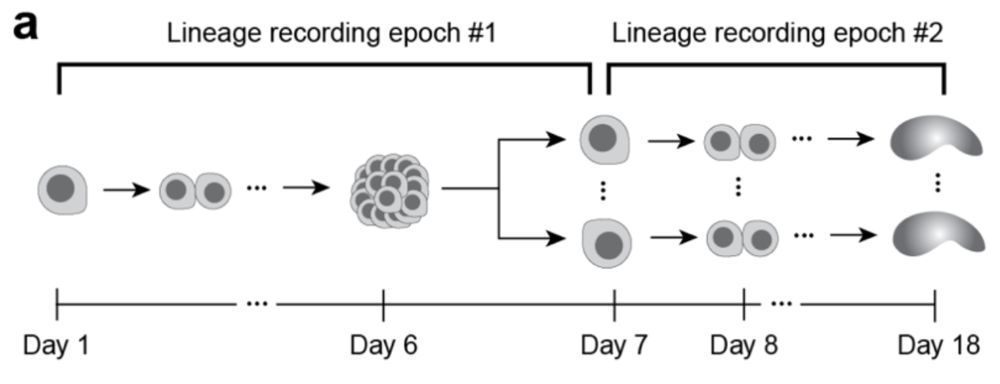
Lineage recording in monoclonal gastruloids reveals heritable modes of early development
Mammalian stem cells possess a remarkable capacity for self-organization, a property that underlies increasingly sophisticated in vitro models of early development. However, even under carefully contr...
www.biorxiv.org
Junhong Choi
@choijunhong.bsky.social
· May 7
Junhong Choi
@choijunhong.bsky.social
· May 7
Junhong Choi
@choijunhong.bsky.social
· May 6

SEARLE SCHOLARS PROGRAM NAMES 15 SCIENTISTS AS SEARLE SCHOLARS FOR 2025
Members of the new class of Searle Scholars pursue ground-breaking research in chemistry and the biomedical sciences. Each receives an award of $300,000 in flexible funding to support his, her, or …
searlescholars.org
Junhong Choi
@choijunhong.bsky.social
· May 6
Junhong Choi
@choijunhong.bsky.social
· May 6
Reposted by Junhong Choi
Naama Aviram
@naamaaviram.bsky.social
· Jan 2
Reposted by Junhong Choi
James Briscoe
@jamesbriscoe.bsky.social
· Dec 19
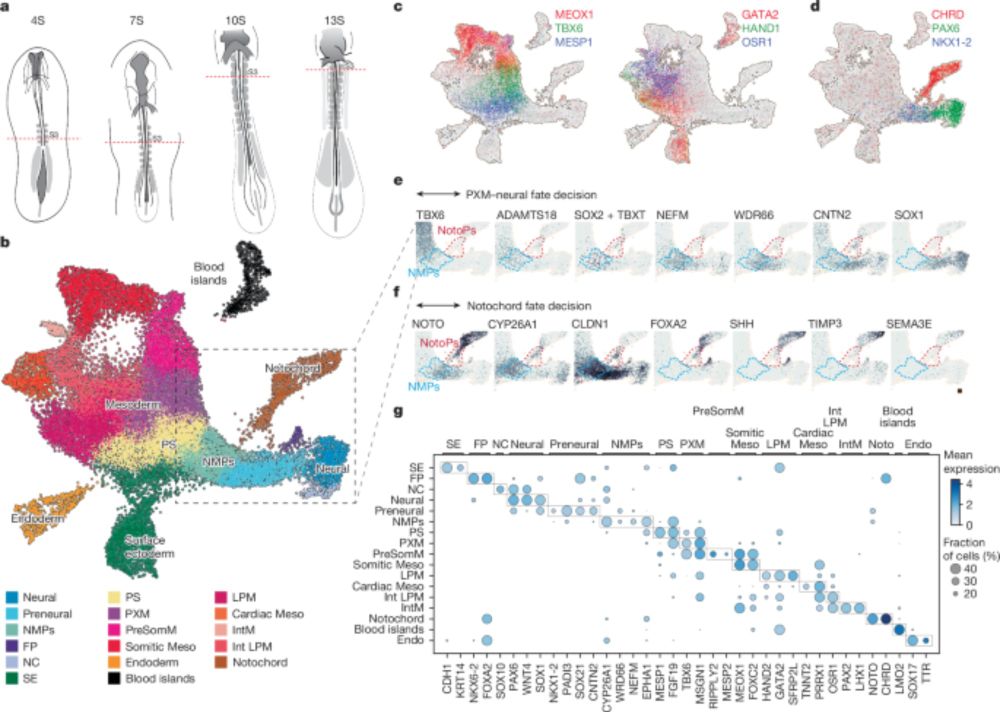
Timely TGFβ signalling inhibition induces notochord - Nature
Through analysis of developing chick embryos and in vitro differentiation of embryonic stem cells, a study develops a method to generate a model of the human trunk with a notochord.
www.nature.com