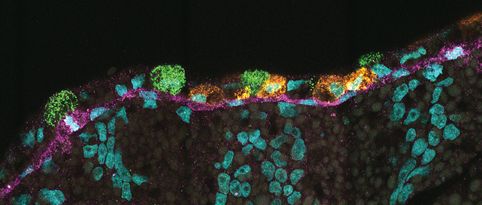
Christian Mayer
@chr-mayer.bsky.social
130 followers
130 following
12 posts
Developmental neuroscientist at the MPI for Biological Intelligence | Gene regulation | Lineage tracing | Single-cell omics | Primarily active on Mastodon (@[email protected]) where I value community-driven exchange science.
Posts
Media
Videos
Starter Packs
Reposted by Christian Mayer
Reposted by Christian Mayer
Reposted by Christian Mayer
Reposted by Christian Mayer
Christian Mayer
@chr-mayer.bsky.social
· May 23
Reposted by Christian Mayer
Reposted by Christian Mayer