@dominik1klein.bsky.social
150 followers
210 following
20 posts
ELLIS PhD student @HelmholtzMunich, Student Researcher @Apple. Interested in ML, Single-Cell Genomics, and People.
Posts
Media
Videos
Starter Packs
Pinned
Reposted
jonas
@josch1.bsky.social
· Apr 18

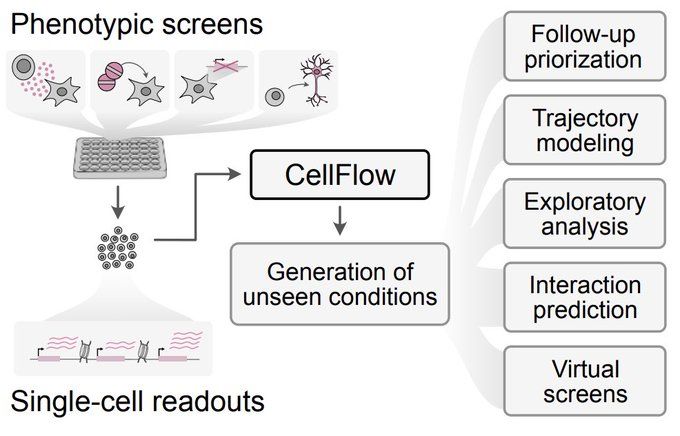
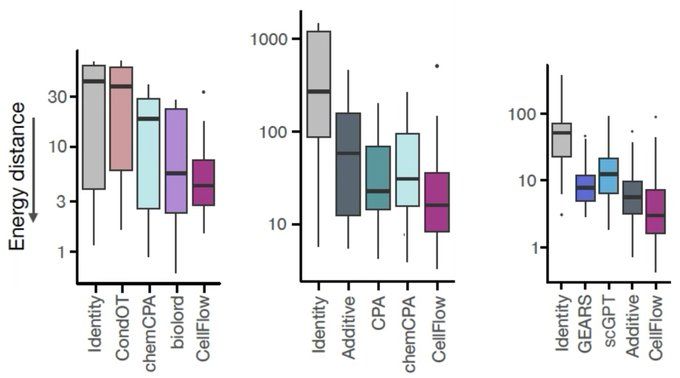
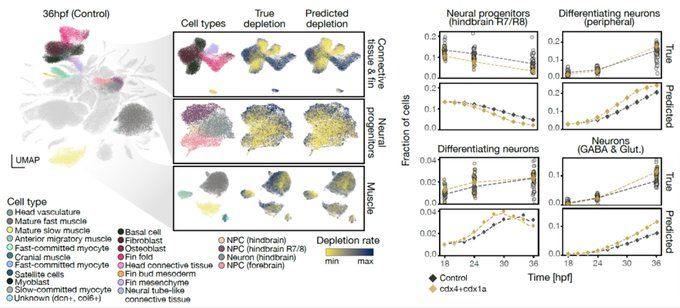
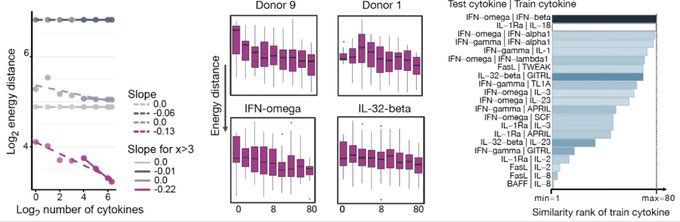
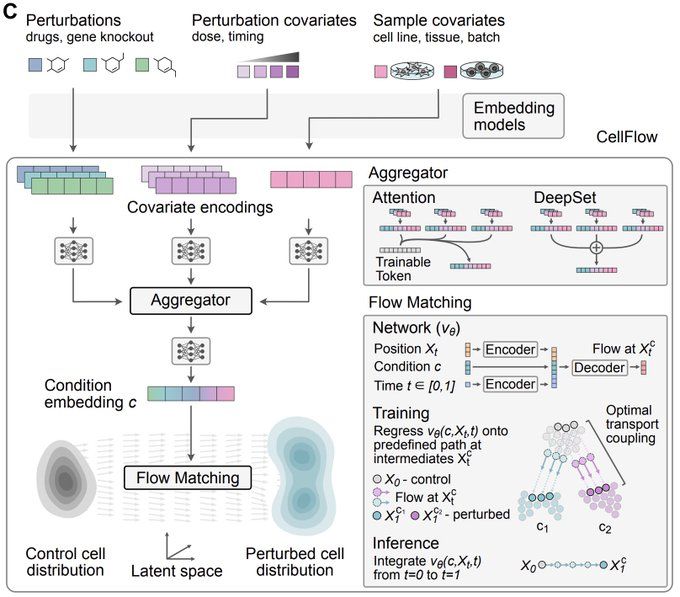
CellFlow enables generative single-cell phenotype modeling with flow matching
High-content phenotypic screens provide a powerful strategy for studying biological systems, but the scale of possible perturbations and cell states makes exhaustive experiments unfeasible. Computatio...
www.biorxiv.org
Reposted
Reposted
Reposted
Nature
@nature.com
· Jan 23
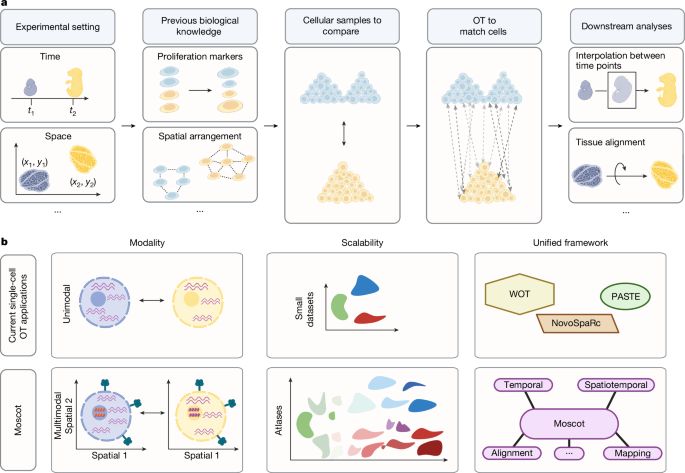
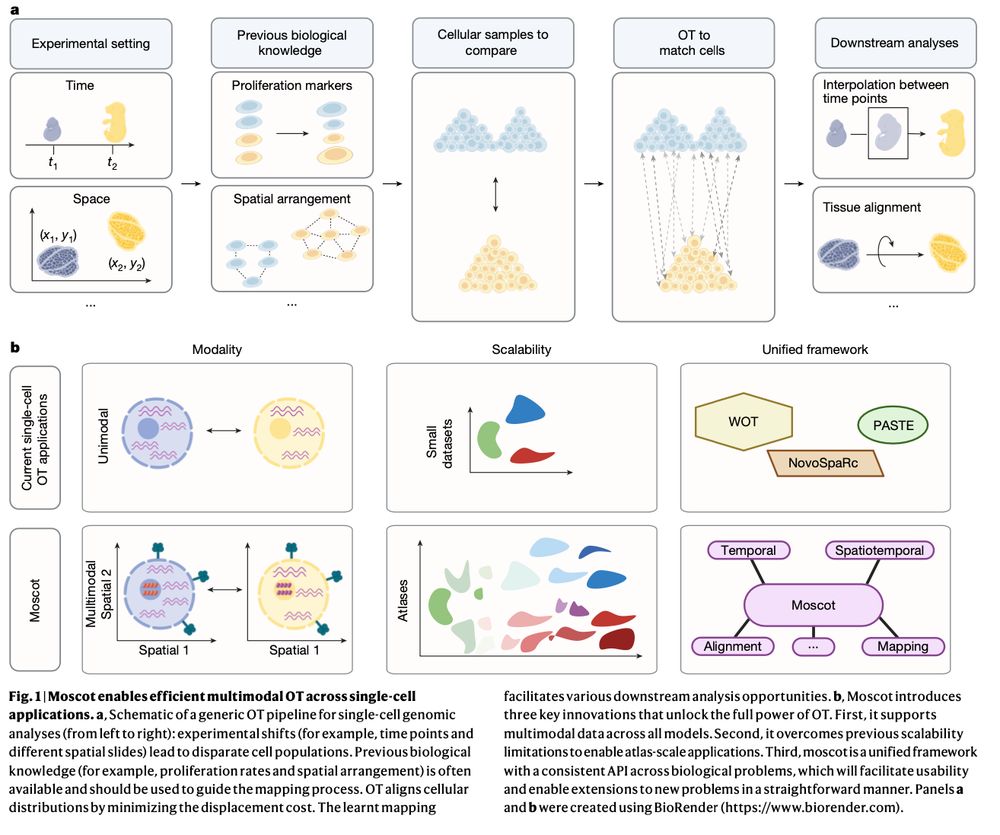
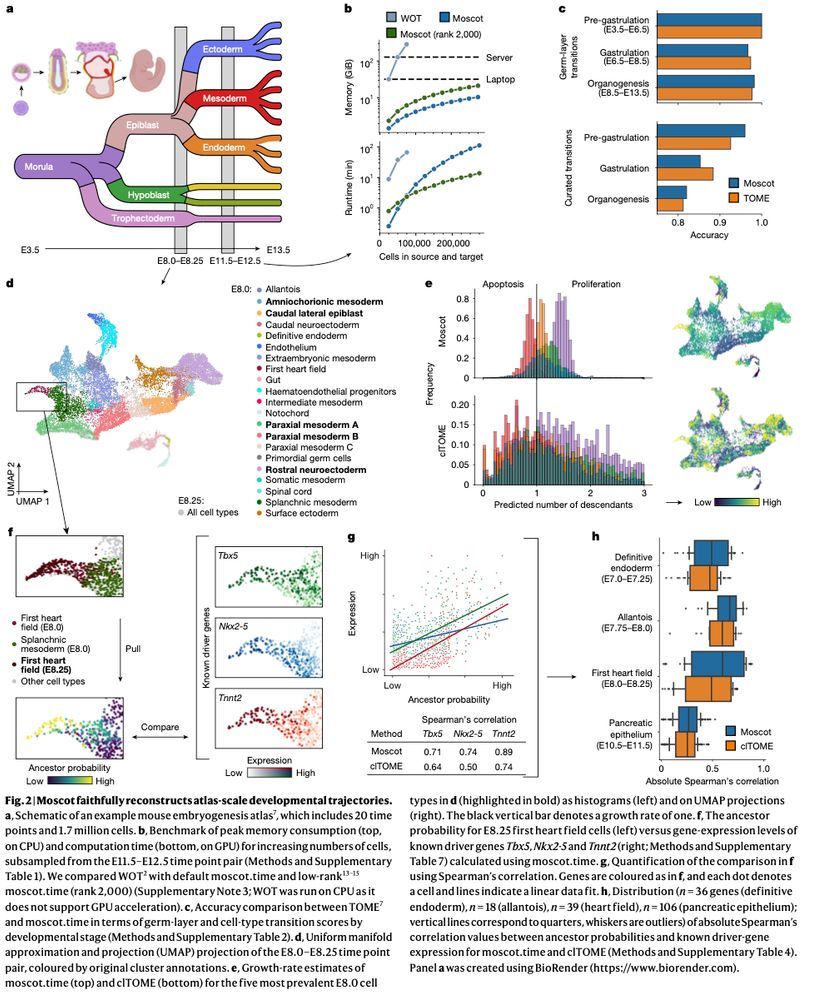
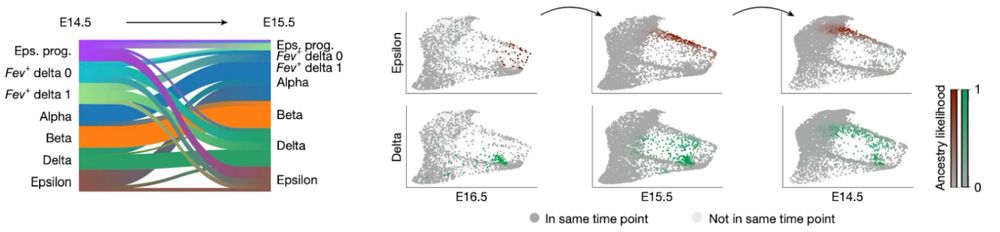
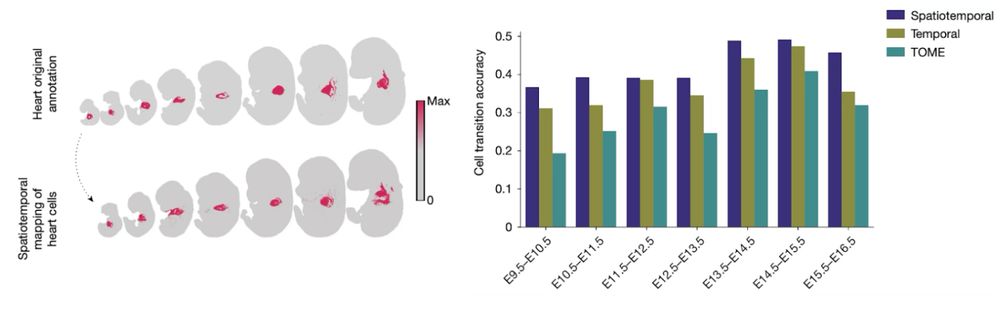
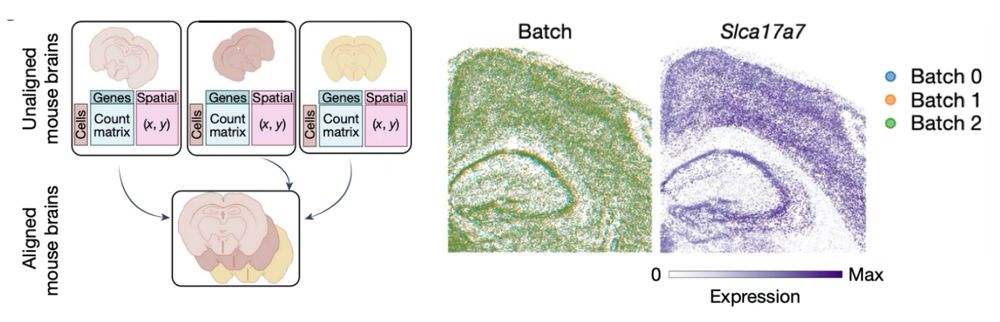
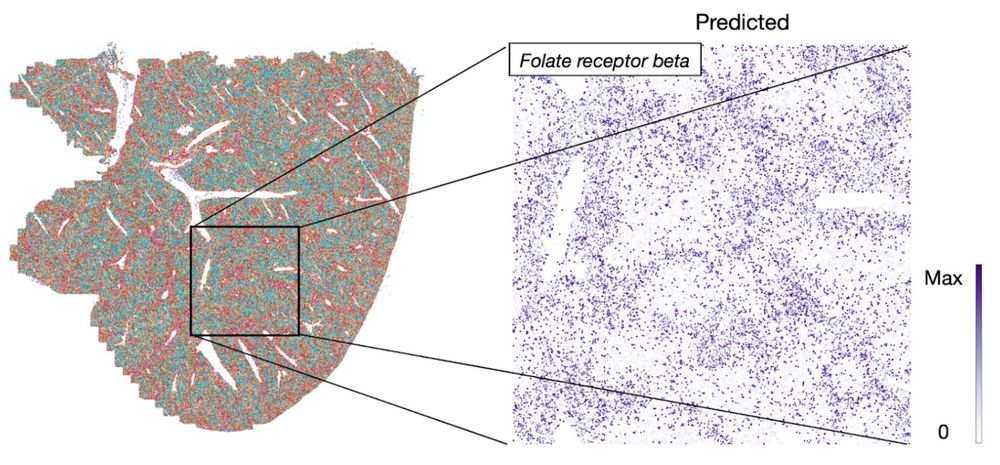
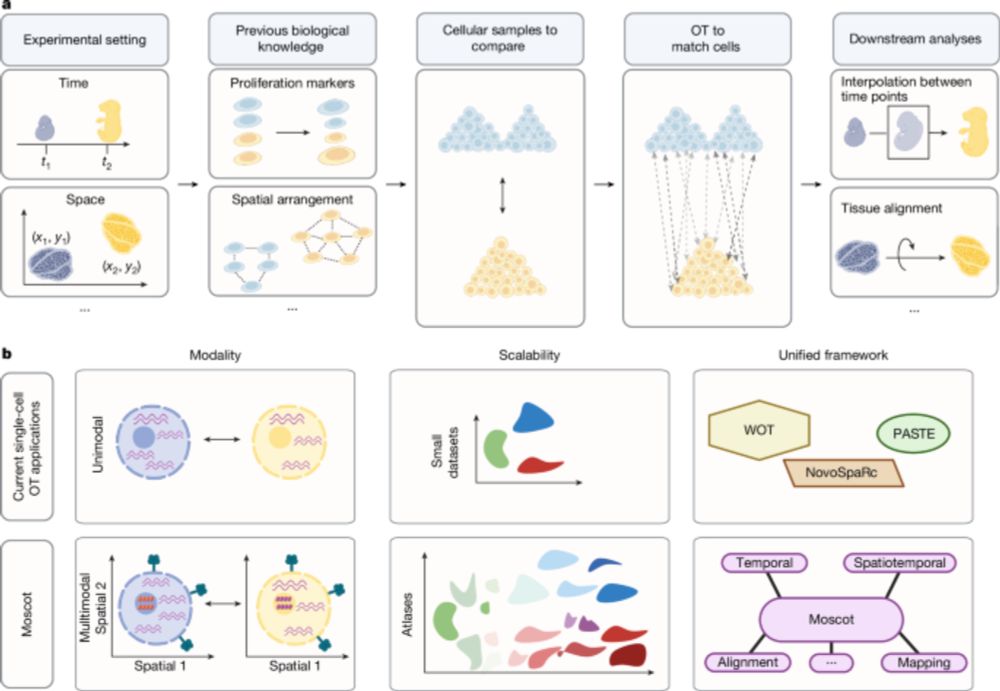
Mapping cells through time and space with moscot - Nature
Moscot is an optimal transport approach that overcomes current limitations of similar methods to enable multimodal, scalable and consistent single-cell analyses of datasets across spatial and temporal dimensions.
go.nature.com
Reposted
Fabian Theis
@fabiantheis.bsky.social
· Jan 22
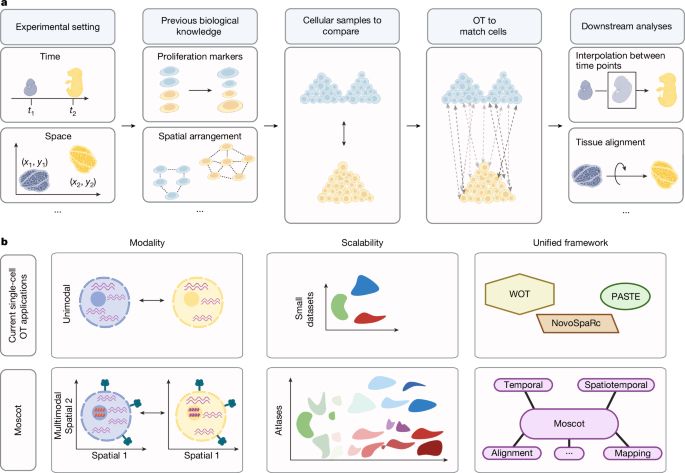
Mapping cells through time and space with moscot - Nature
Moscot is an optimal transport approach that overcomes current limitations of similar methods to enable multimodal, scalable and consistent single-cell analyses of datasets across spatial and temporal...
www.nature.com