jonas
@josch1.bsky.social
130 followers
200 following
14 posts
dev bio. ML. organoids. single cells @IHB
Posts
Media
Videos
Starter Packs
Pinned
jonas
@josch1.bsky.social
· Apr 18

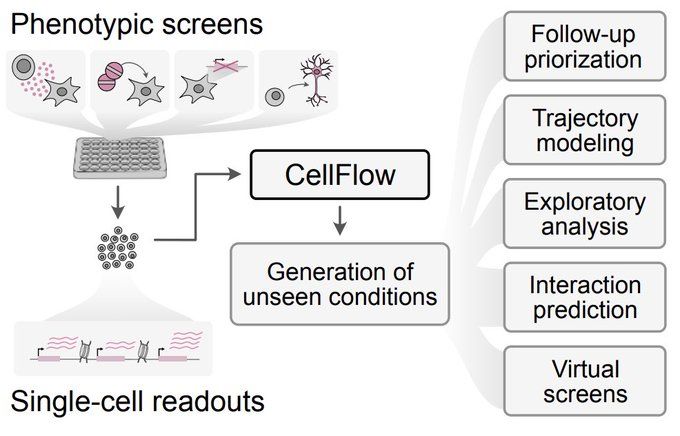
CellFlow enables generative single-cell phenotype modeling with flow matching
High-content phenotypic screens provide a powerful strategy for studying biological systems, but the scale of possible perturbations and cell states makes exhaustive experiments unfeasible. Computatio...
www.biorxiv.org
Reposted by jonas
Anshul Kundaje
@anshulkundaje.bsky.social
· Jul 10
Reposted by jonas
Anshul Kundaje
@anshulkundaje.bsky.social
· Jun 28
jonas
@josch1.bsky.social
· Jun 24
jonas
@josch1.bsky.social
· Jun 24
jonas
@josch1.bsky.social
· Jun 24
Reposted by jonas
Quan Xu
@quanxu.bsky.social
· May 13
Camp Lab
@graycamplab.bsky.social
· May 12

An integrated transcriptomic cell atlas of human endoderm-derived organoids - Nature Genetics
The human endoderm-derived organoid cell atlas (HEOCA) presents an integrative analysis of single-cell transcriptomes across different conditions, sources and protocols. It compares cell types and sta...
nature.com
Reposted by jonas
Reposted by jonas
Reposted by jonas
Reposted by jonas
jonas
@josch1.bsky.social
· Apr 24
jonas
@josch1.bsky.social
· Apr 24
jonas
@josch1.bsky.social
· Apr 18

CellFlow enables generative single-cell phenotype modeling with flow matching
High-content phenotypic screens provide a powerful strategy for studying biological systems, but the scale of possible perturbations and cell states makes exhaustive experiments unfeasible. Computatio...
www.biorxiv.org
Reposted by jonas
Reposted by jonas
Fabian Theis
@fabiantheis.bsky.social
· Jan 22
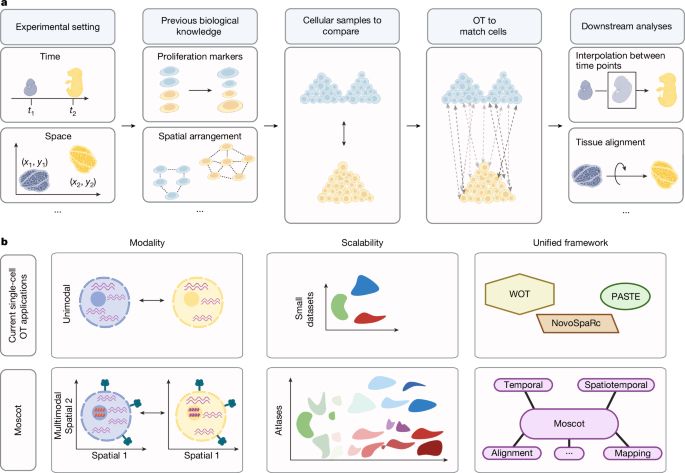
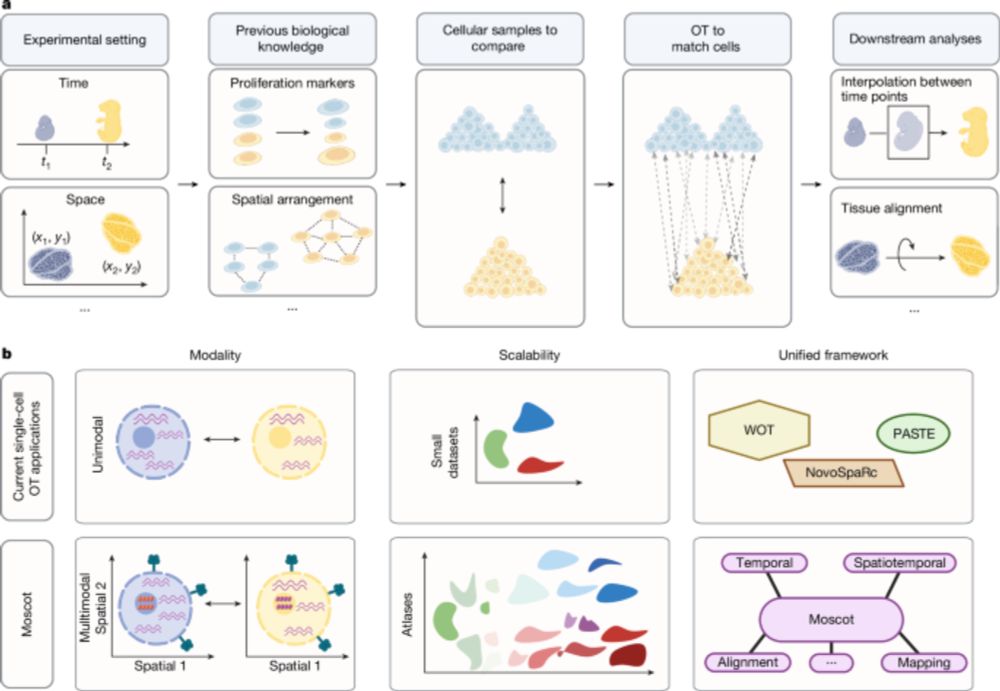
Mapping cells through time and space with moscot - Nature
Moscot is an optimal transport approach that overcomes current limitations of similar methods to enable multimodal, scalable and consistent single-cell analyses of datasets across spatial and temporal...
www.nature.com
Reposted by jonas
Hank Green
@hankgreen.bsky.social
· Dec 28
Reposted by jonas
Leander
@le-and-er.bsky.social
· Dec 19

Meinung: »Human Cell Atlas«: Das wird die wichtigste Wissenschaft des 21. Jahrhunderts - Kolumne
Gerade ist ein ganzes Bündel Publikationen aus einem einzigen Forschungsprojekt erschienen. Sie weisen in die Zukunft einer neuen Wissenschaft: Lernende Maschinen helfen jetzt, die Maschinerie des Leb...
www.spiegel.de