Leander
@le-and-er.bsky.social
240 followers
140 following
15 posts
Postdoc between Helmholtz Munich and ETH Zurich | Theis Lab & Treutlein Lab | Computational Biology, Machine Learning & Organoids
Posts
Media
Videos
Starter Packs
Reposted by Leander
Reposted by Leander
Leander
@le-and-er.bsky.social
· Feb 21
Reposted by Leander
Cedric Boeckx
@cedricboeckx.bsky.social
· Feb 15
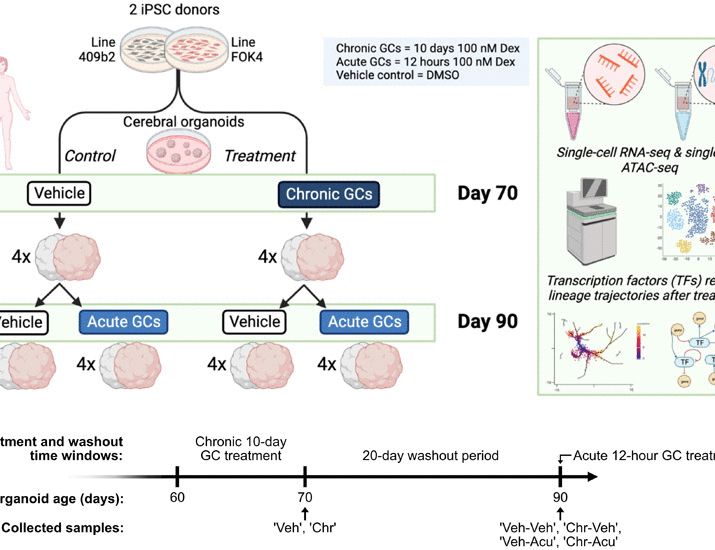
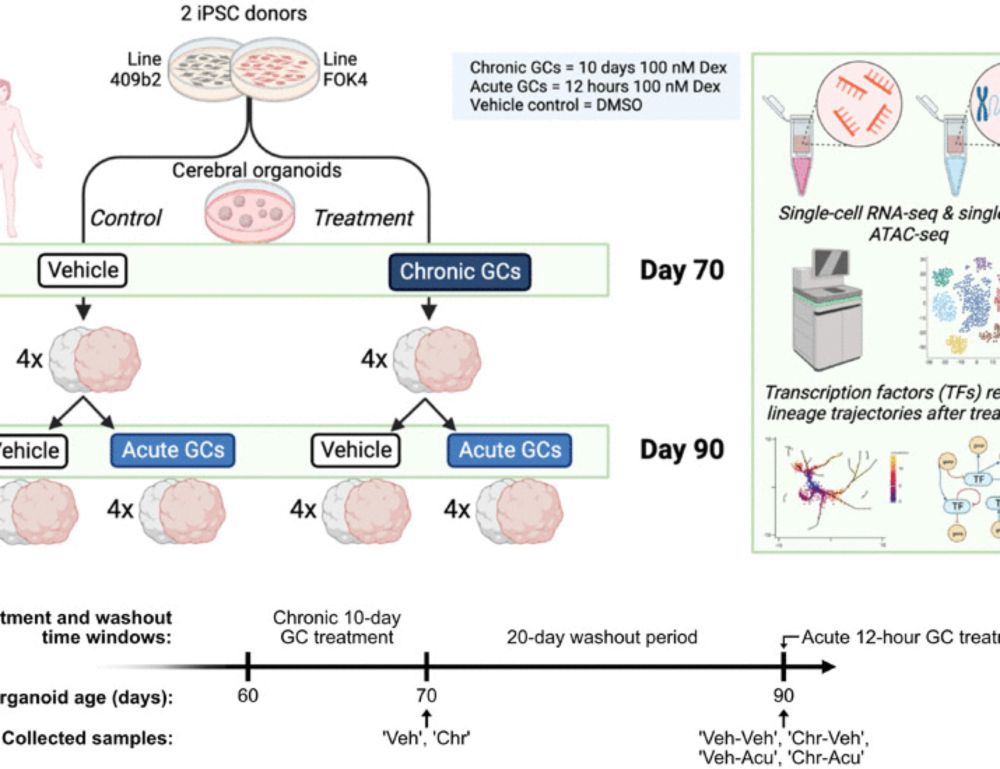
Chronic exposure to glucocorticoids amplifies inhibitory neuron cell fate during human neurodevelopment in organoids
Chronic exposure to glucocorticoids during brain development leads to priming of the inhibitory neuron lineages in organoids.
www.science.org
Leander
@le-and-er.bsky.social
· Dec 19

Meinung: »Human Cell Atlas«: Das wird die wichtigste Wissenschaft des 21. Jahrhunderts - Kolumne
Gerade ist ein ganzes Bündel Publikationen aus einem einzigen Forschungsprojekt erschienen. Sie weisen in die Zukunft einer neuen Wissenschaft: Lernende Maschinen helfen jetzt, die Maschinerie des Leb...
www.spiegel.de
Leander
@le-and-er.bsky.social
· Nov 21

The Human Cell Atlas: towards a first draft atlas
In a collection of research articles and related content, the Human Cell Atlas consortium presents tools, data and ideas towards the generation of their first draft atlas of cells in the human body.
www.nature.com
Leander
@le-and-er.bsky.social
· Nov 21
Leander
@le-and-er.bsky.social
· Nov 21
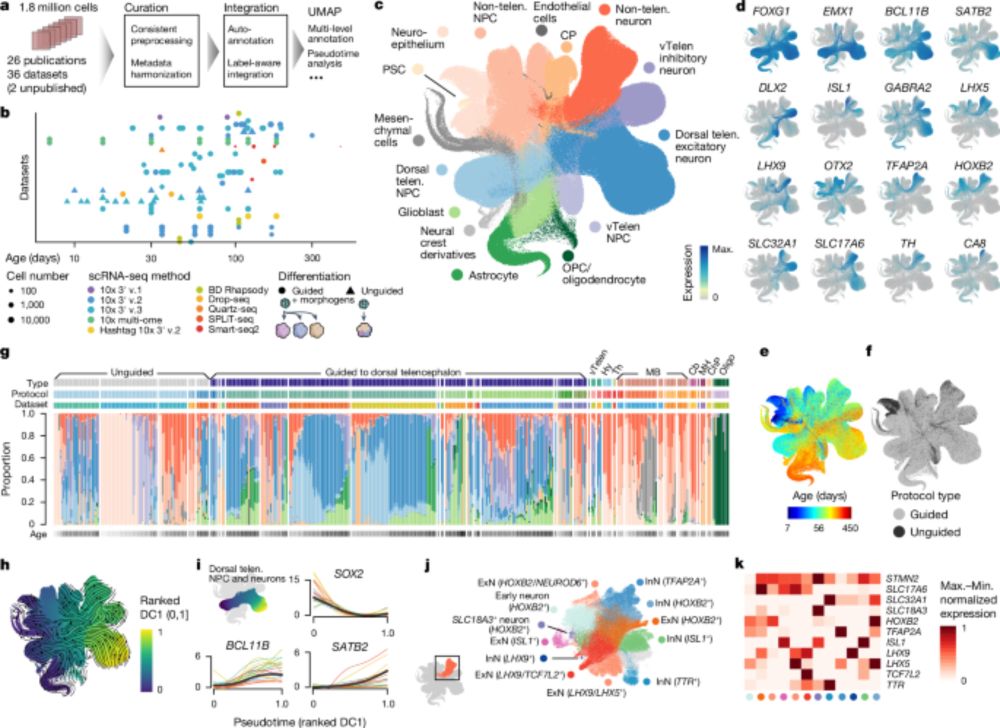
An integrated transcriptomic cell atlas of human neural organoids - Nature
A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, sho...
www.nature.com