Dongwook Kim
@dongwookkim.bsky.social
160 followers
92 following
4 posts
Developing fast and easy methods for #phylogenetics and #bioinformatics | PhD in Bioinformatics | Postdoc @ Comparative Genomics Lab, UNIL/SIB🇨🇭| Formerly @ Steinegger Lab, SNU🇰🇷 | he/him
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Dongwook Kim
George Bouras
@gbouras13.bsky.social
· Aug 8

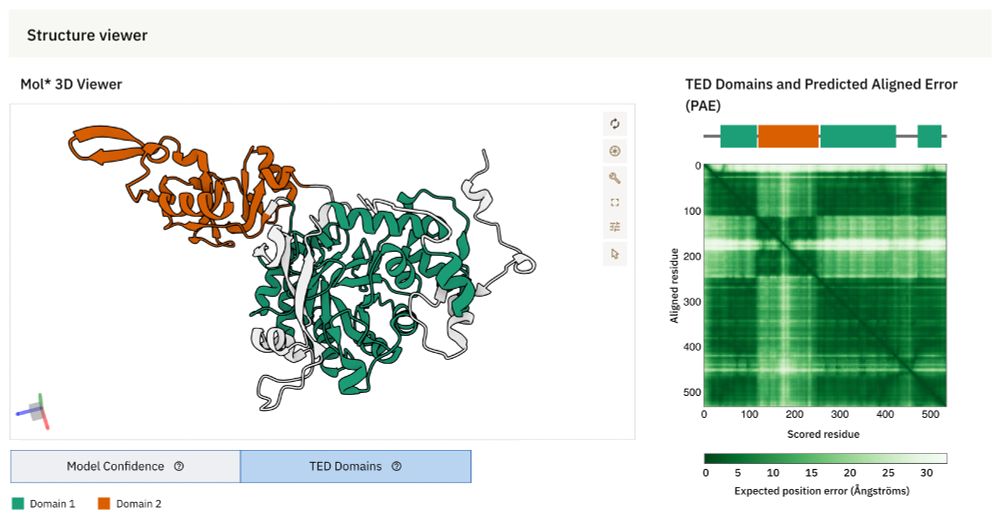
Protein Structure Informed Bacteriophage Genome Annotation with Phold
Bacteriophage (phage) genome annotation is essential for understanding their functional potential and suitability for use as therapeutic agents. Here we introduce Phold, an annotation framework utilis...
www.biorxiv.org
Reposted by Dongwook Kim
Chan Yeong Kim
@chanyeong-kim.bsky.social
· Jul 21

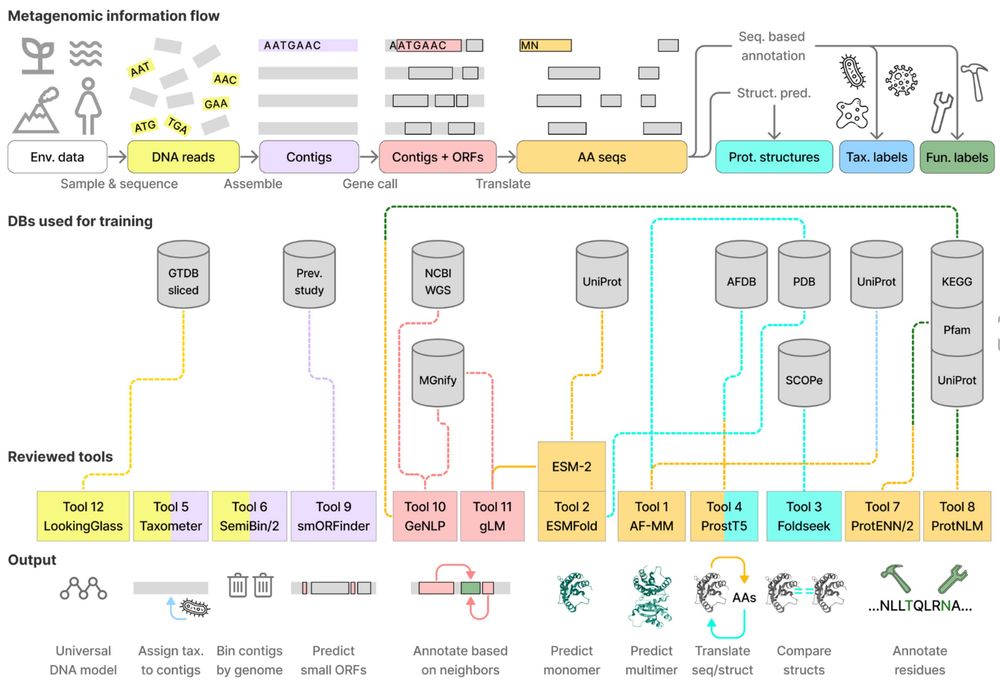
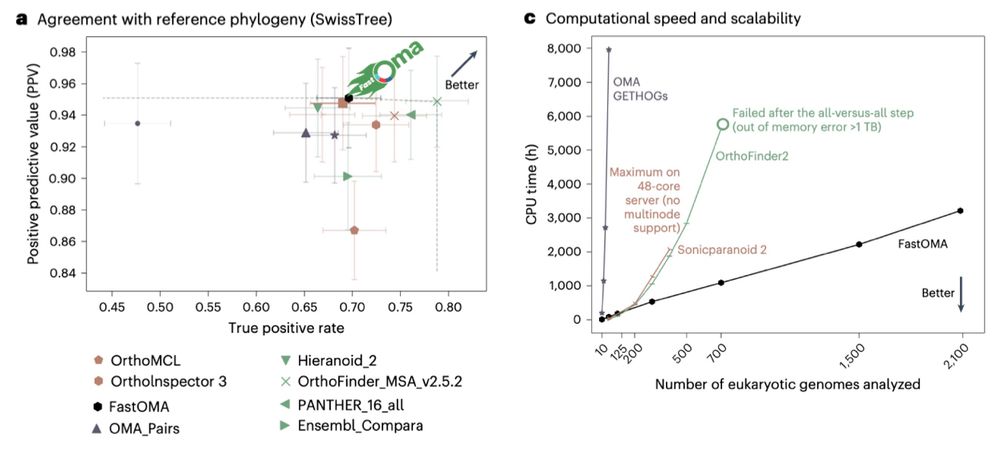
Planetary microbiome structure and generalist-driven gene flow across disparate habitats
Microbes are ubiquitous on Earth, forming microbiomes that sustain macroscopic life and biogeochemical cycles. Microbial dispersion, driven by natural processes and human activities, interconnects mic...
www.biorxiv.org
Reposted by Dongwook Kim
Laurie Belcher
@lauriebelch.bsky.social
· Jul 16
Reposted by Dongwook Kim
Dongwook Kim
@dongwookkim.bsky.social
· Jun 3
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
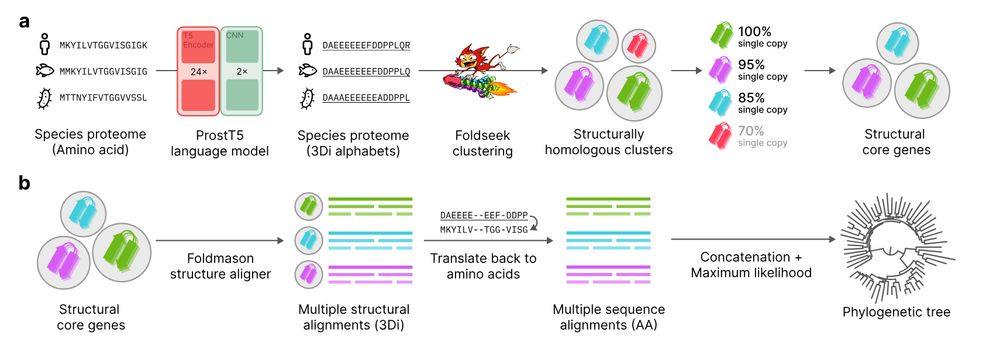
Christophe Dessimoz
@dessimoz.bsky.social
· Feb 26
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim
Reposted by Dongwook Kim