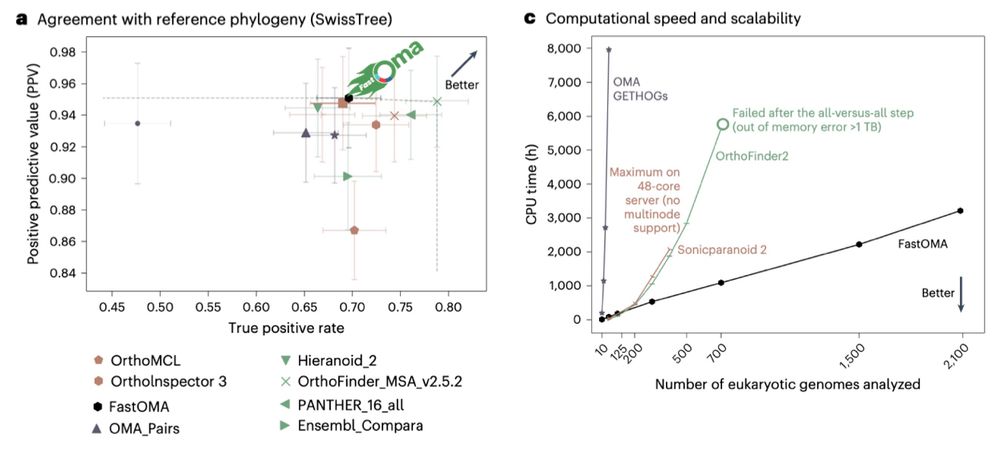
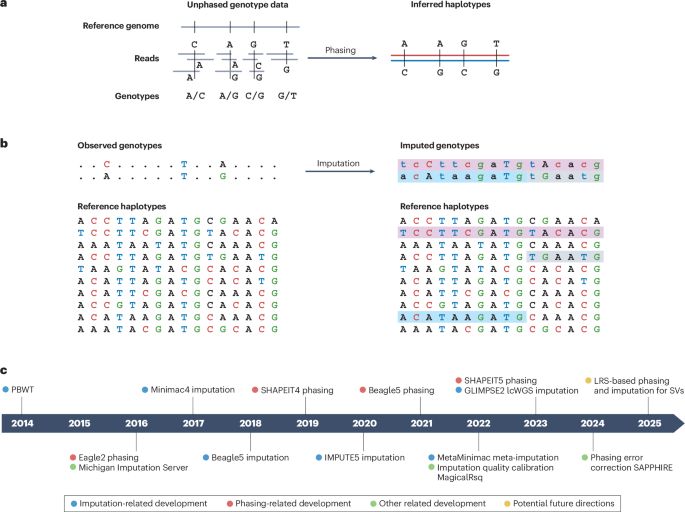
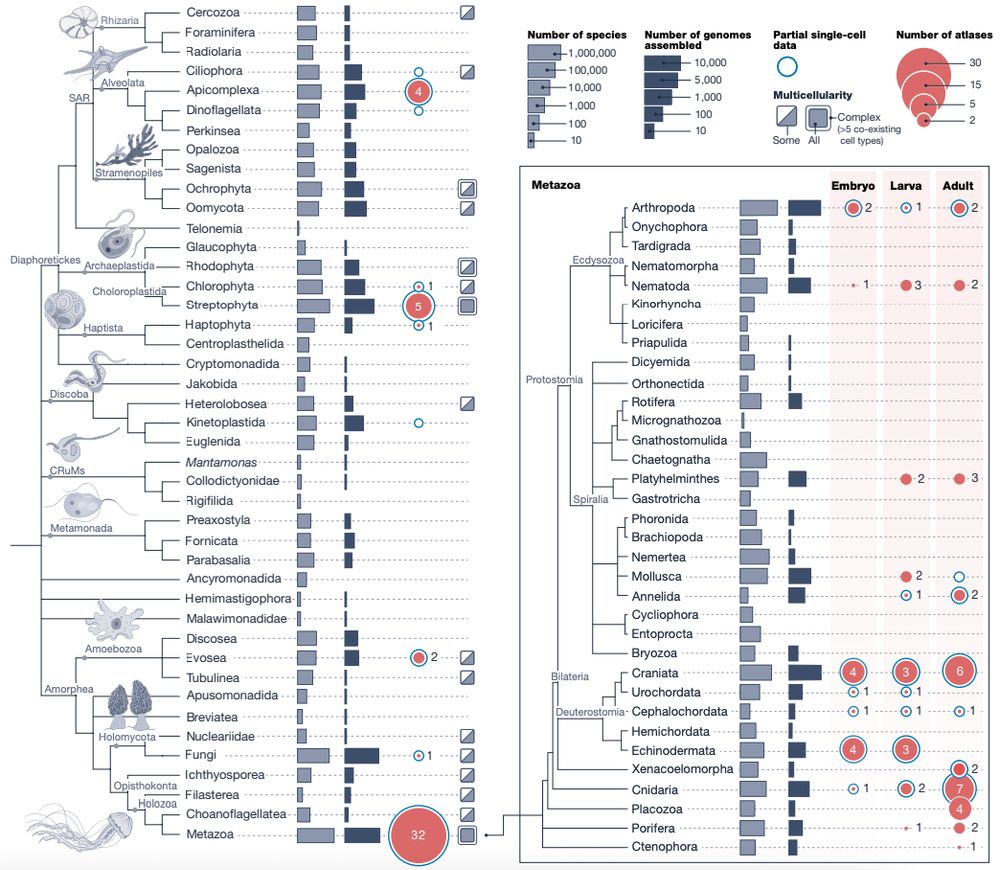
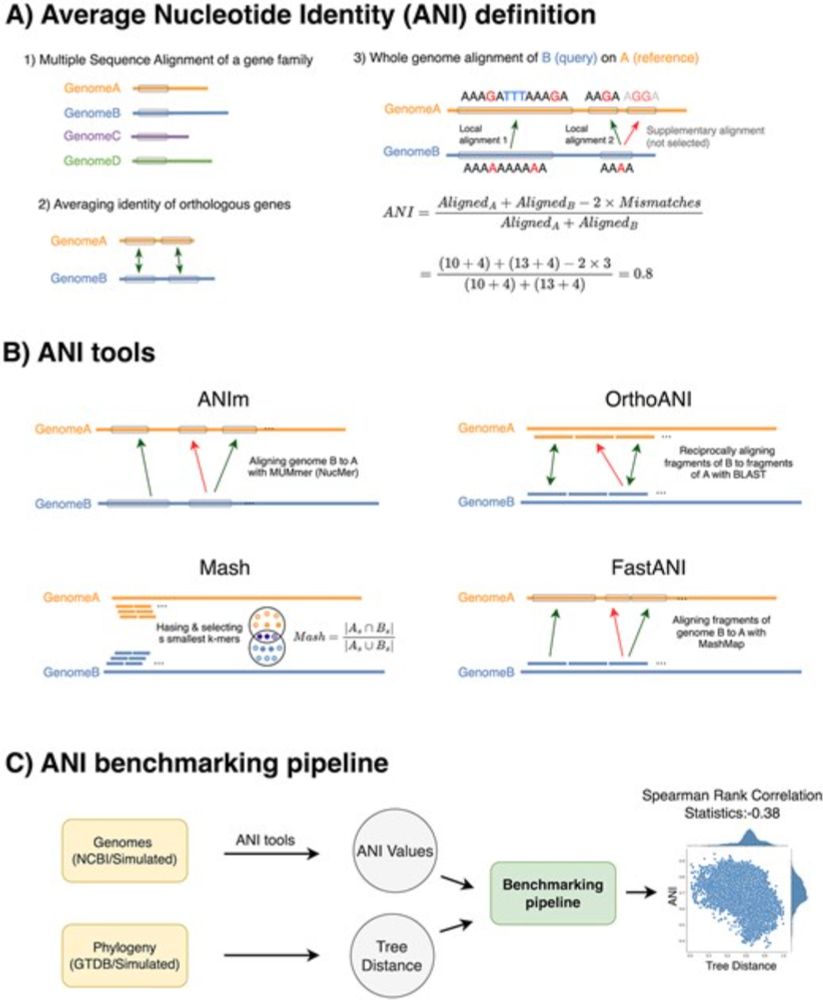
Sina Majidian
@sinamajidian.bsky.social
1.3K followers
1.6K following
89 posts
On the academic job market | How are species compared to one another across different genomic regions? Postdoc at Langmead Lab, Johns Hopkins | Comparative #genomics at scale | Formerly at UNIL/SIB/WUR | sinamajidian.github.io
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian
Reposted by Sina Majidian