Xian Chang
@xian-chang.bsky.social
160 followers
140 following
9 posts
Bioinformatics postdoc at INRAE in Toulouse, France.
Working on data structures and algorithms for pangenome graphs
Posts
Media
Videos
Starter Packs
Reposted by Xian Chang
Reposted by Xian Chang
Xian Chang
@xian-chang.bsky.social
· Mar 22
Xian Chang
@xian-chang.bsky.social
· Mar 21
Reposted by Xian Chang
Jouni Sirén
@jltsiren.bsky.social
· Mar 4
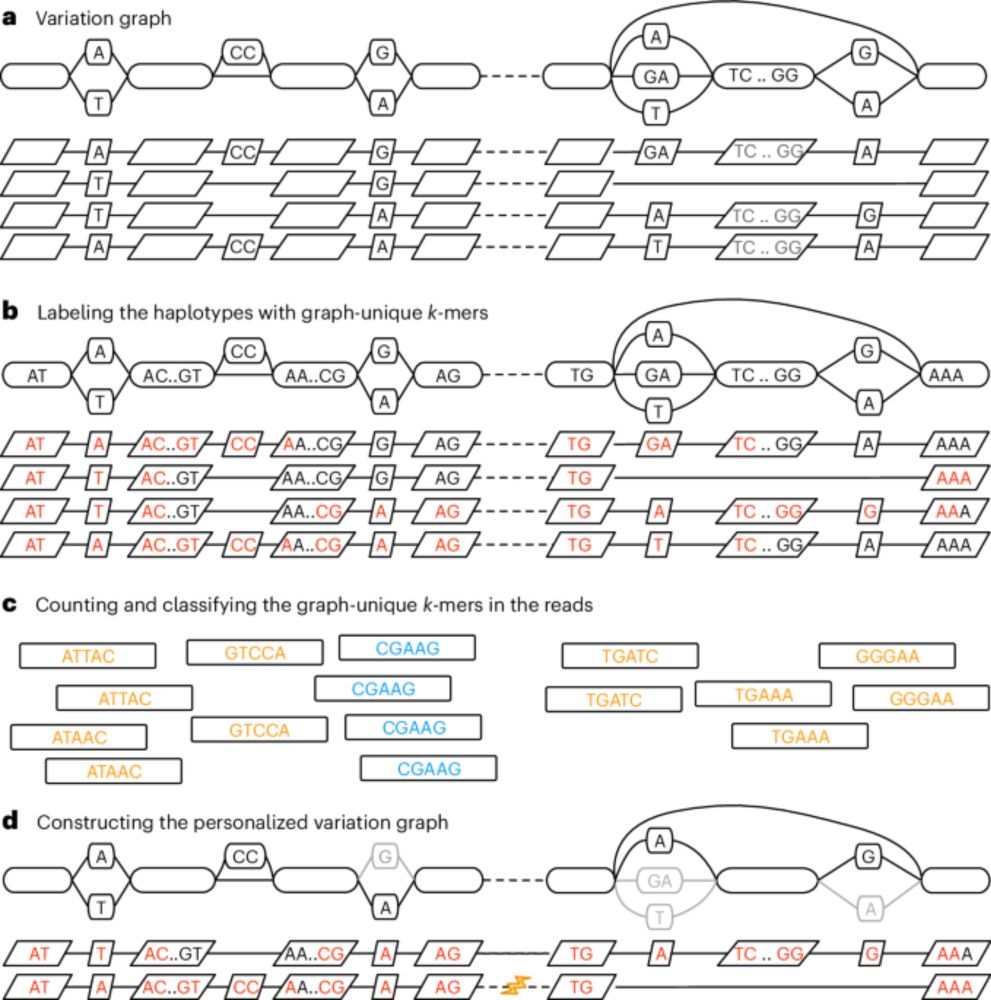
Personalized pangenome references - Nature Methods
This work introduces a k-mer-based approach to customizing a pangenome reference, making it more relevant to a new sample of interest. This method enhances the accuracy of genotyping small variants an...
www.nature.com
Reposted by Xian Chang
Reposted by Xian Chang