Dubcovsky Lab
@dubcovskylab.bsky.social
110 followers
24 following
14 posts
Updates from Jorge Dubcovsky's lab at UC Davis
Account run by students and postdocs
https://dubcovskylab.ucdavis.edu/home
Posts
Media
Videos
Starter Packs
Dubcovsky Lab
@dubcovskylab.bsky.social
· Aug 13
Dubcovsky Lab
@dubcovskylab.bsky.social
· Aug 13

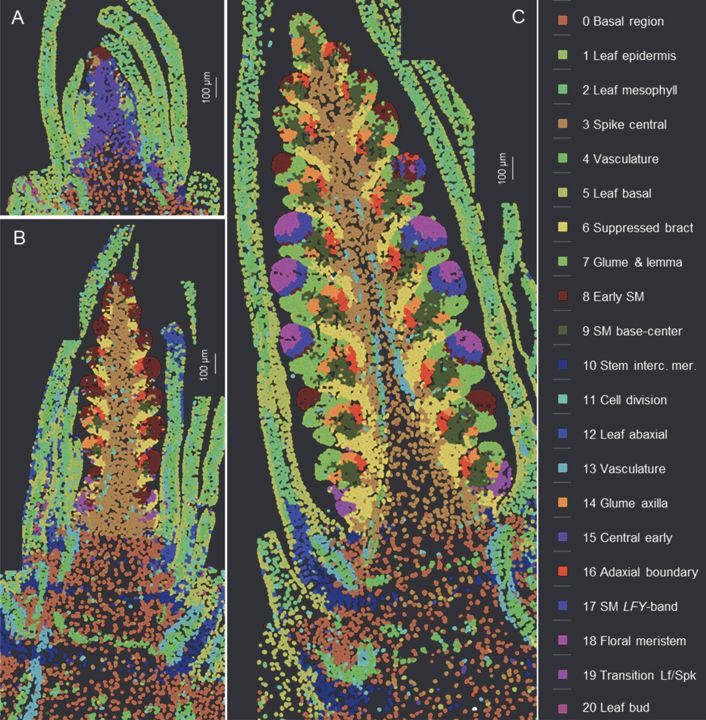
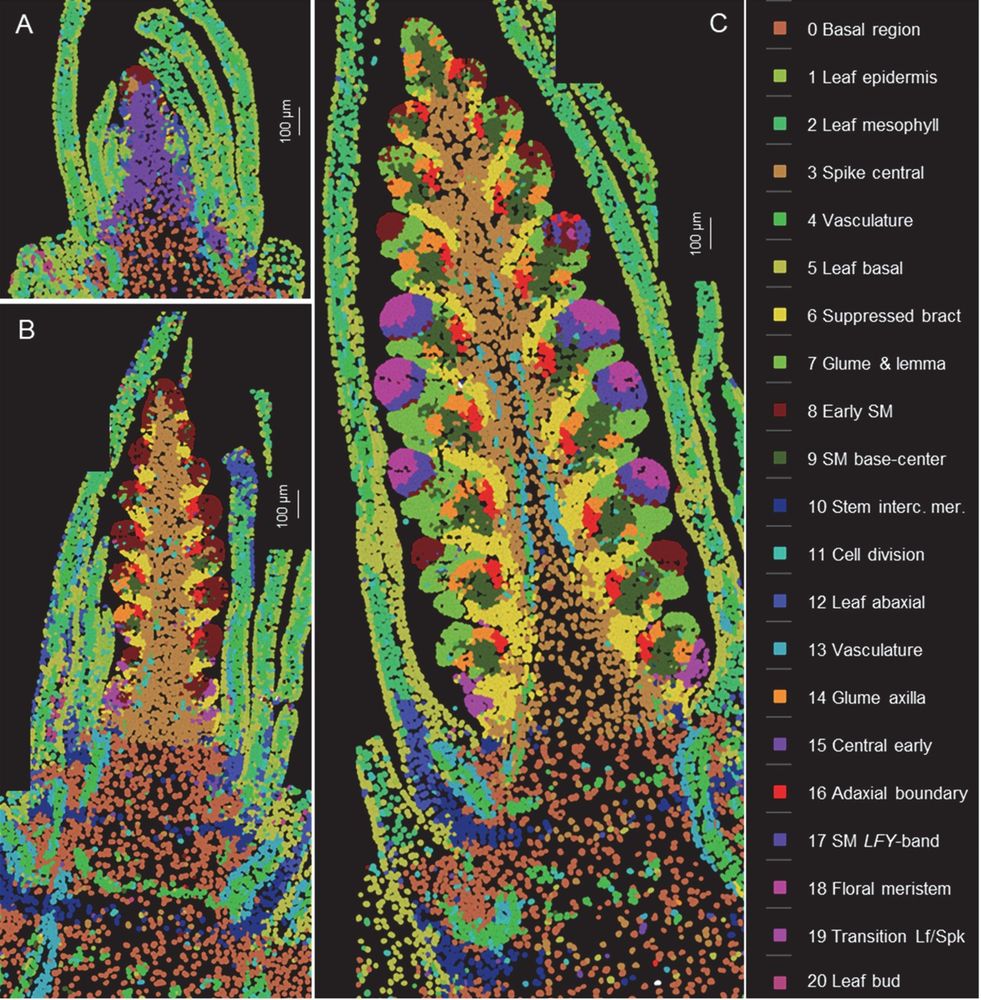
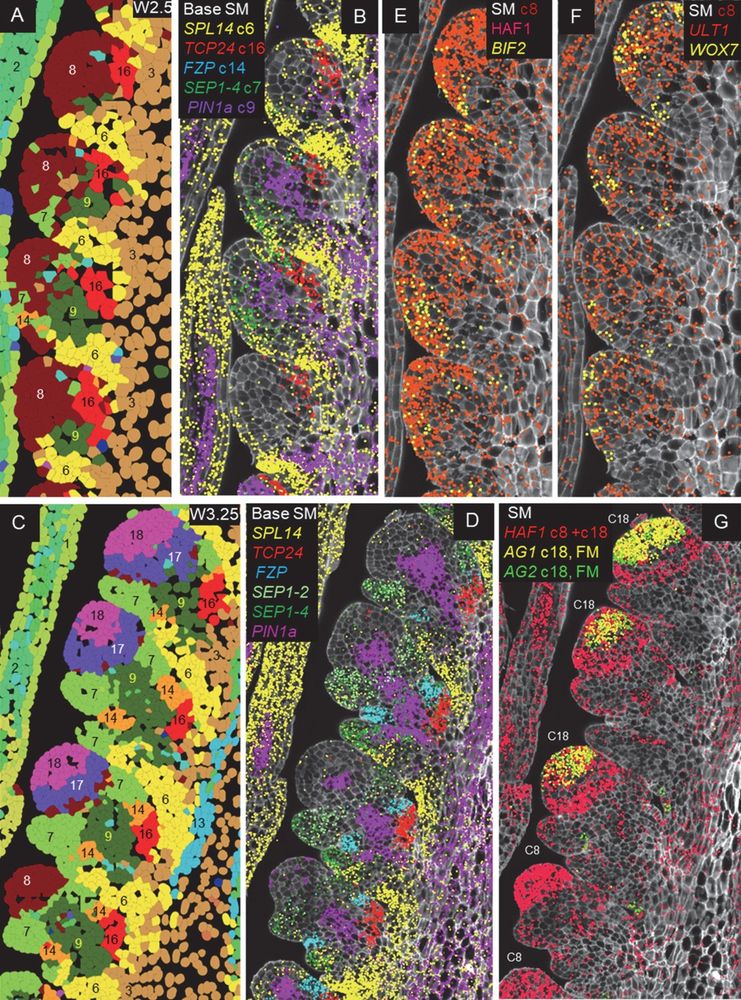
Spatial and single-cell expression analyses reveal complex expression domains in early wheat spike development
Wheat is important for global food security and understanding the molecular mechanisms driving spike and spikelet development can inform the development of more productive varieties. In this study, we...
www.biorxiv.org
Reposted by Dubcovsky Lab
UCDavisPlants
@ucdavisplants.bsky.social
· Aug 12

Durum wheat with Yr78 and common wheat with Yr78 and Yr36 in coupling show enhanced stripe rust resistance
Introgression of Yr78 (where Yr is yellow rust) from common wheat into durum wheat enhances resistance to stripe rust in tetraploid wheat. A combination of Yr36 and Yr78 resistance genes in coupling...
acsess.onlinelibrary.wiley.com
Dubcovsky Lab
@dubcovskylab.bsky.social
· Aug 10
Dubcovsky Lab
@dubcovskylab.bsky.social
· Aug 10

Durum wheat with Yr78 and common wheat with Yr78 and Yr36 in coupling show enhanced stripe rust resistance
Introgression of Yr78 (where Yr is yellow rust) from common wheat into durum wheat enhances resistance to stripe rust in tetraploid wheat. A combination of Yr36 and Yr78 resistance genes in coupling...
acsess.onlinelibrary.wiley.com
Reposted by Dubcovsky Lab
Reposted by Dubcovsky Lab
Reposted by Dubcovsky Lab
Dubcovsky Lab
@dubcovskylab.bsky.social
· Mar 14

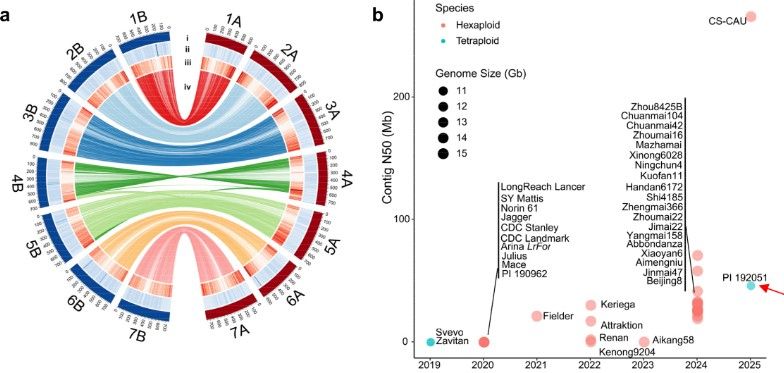
Blocking the scissors: New genetic tool makes grain breeding faster
Scientists have developed a new genetic tool that makes it easier and faster to breed grains that grow more efficiently and are better adapted to different environments. They're using a ground-breakin...
www.plantsciences.ucdavis.edu
Reposted by Dubcovsky Lab
Plantae.org
@plantaeofficial.bsky.social
· Mar 14
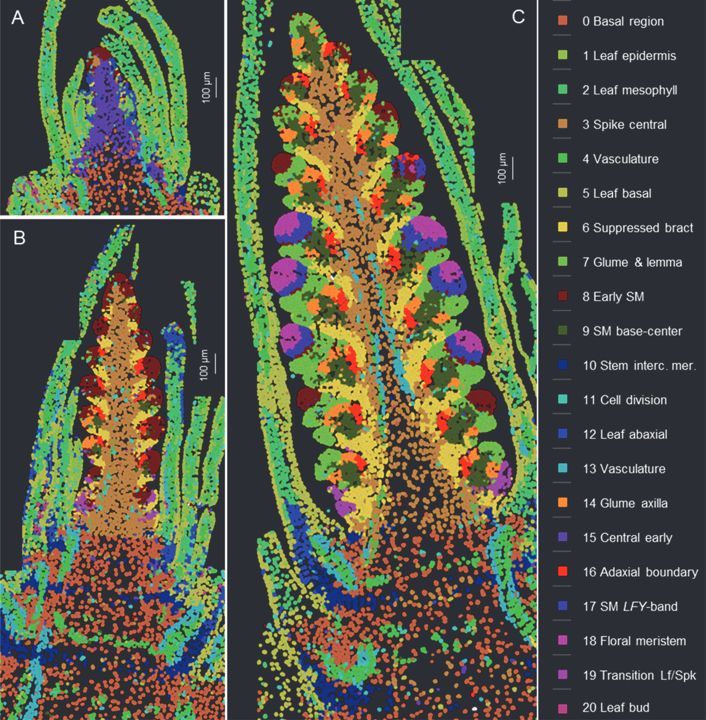
Single cell analysis of wheat spike development | Plantae
This preprint wins the award for “most beautiful paper” this week. Xu, Lin et al. carried out an expression analysis of developing wheat spikes at three developmental stages, using both single-cell…
buff.ly
Reposted by Dubcovsky Lab
Reposted by Dubcovsky Lab
UCDavisPlants
@ucdavisplants.bsky.social
· Feb 18

A triplicated wheat-rye chromosome segment including several 12-OXOPHYTODIENOATE REDUCTASE III genes influences magnesium partitioning and impacts wheat performance at low magnesium supply
We previously reported a structural rearrangement between wheat (Triticum aestivum) and rye (Secale cereale) chromosomes 1BS/1RS that increased the do…
www.sciencedirect.com
Dubcovsky Lab
@dubcovskylab.bsky.social
· Feb 14