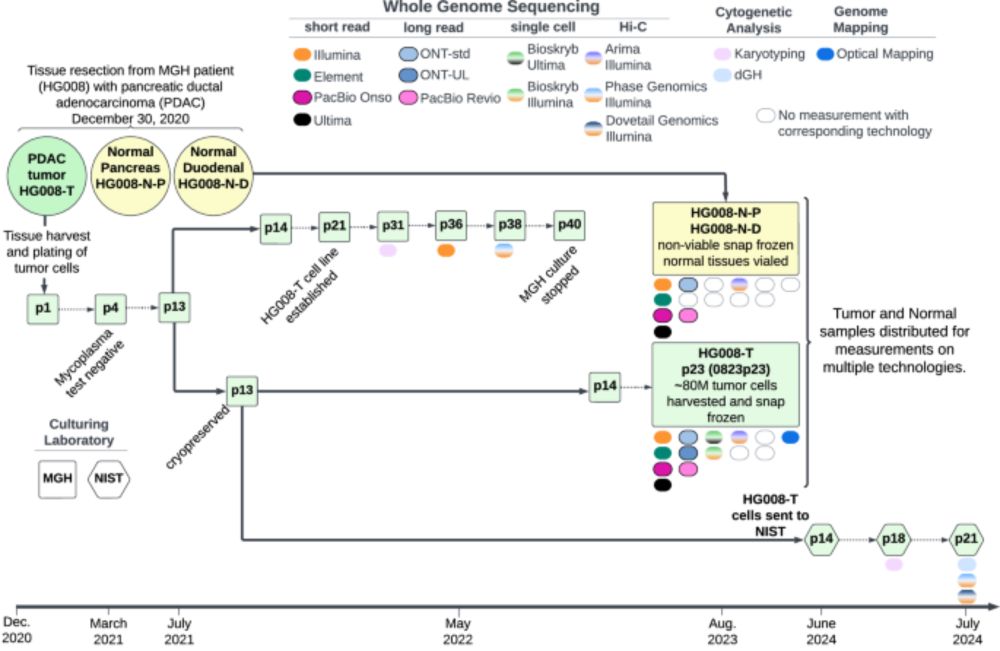
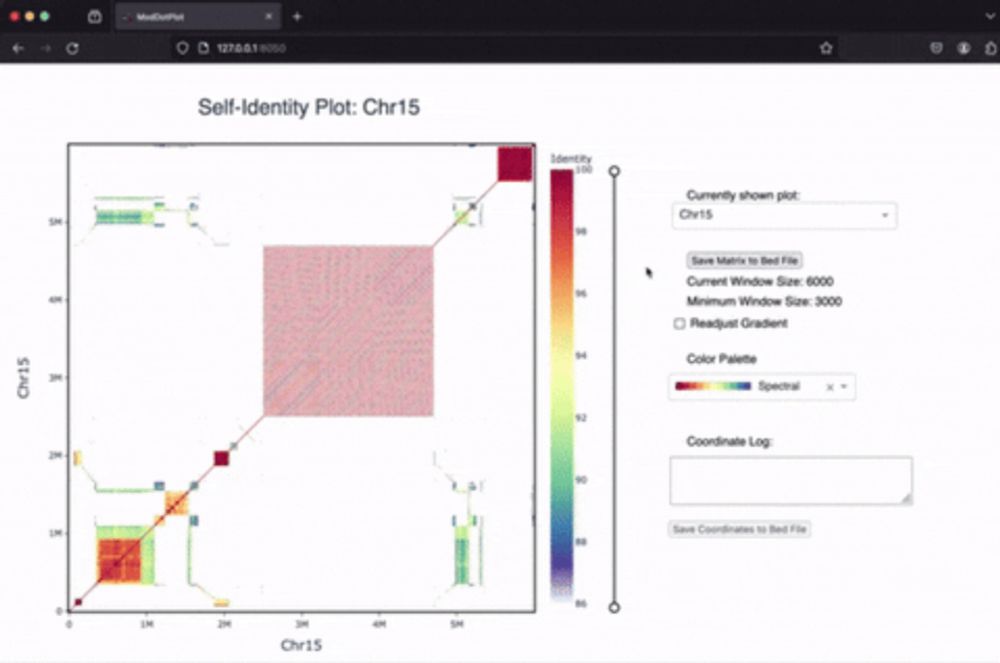
Genome in a Bottle Consortium
@genomeinabottle.bsky.social
450 followers
61 following
6 posts
The Genome in a Bottle Consortium develops reference materials, reference data, and reference methods needed to benchmark human genome sequencing
Posts
Media
Videos
Starter Packs
Reposted by Genome in a Bottle Consortium
Reposted by Genome in a Bottle Consortium
resgen.io
@resgen-io.bsky.social
· Feb 4
Reposted by Genome in a Bottle Consortium
Reposted by Genome in a Bottle Consortium
Reposted by Genome in a Bottle Consortium
Corey Watson
@cdubsig.bsky.social
· Nov 1