Gräter lab
@graeterlab.bsky.social
310 followers
46 following
88 posts
We aim at unraveling the intricacies of molecular perception and designing sensing molecular systems from the ground up. We work at the interface of biophysics, biochemistry, and materials science.
@ Max Planck Institute for Polymer Research, Mainz.
Posts
Media
Videos
Starter Packs
Reposted by Gräter lab
Reposted by Gräter lab
Reposted by Gräter lab
Gräter lab
@graeterlab.bsky.social
· May 19
Gräter lab
@graeterlab.bsky.social
· May 9
Gräter lab
@graeterlab.bsky.social
· Apr 4

Max Planck Research Group Leader (W2) in Molecular Design
We are looking for exceptional early-career scientists conducting computational research with a proven record of accomplishment. The primary focus of this call is on candidates proposing research on b...
tinyurl.com
Reposted by Gräter lab
Gräter lab
@graeterlab.bsky.social
· Apr 28
Reposted by Gräter lab
Gräter lab
@graeterlab.bsky.social
· Apr 8
Gräter lab
@graeterlab.bsky.social
· Apr 4

Max Planck Research Group Leader (W2) in Molecular Design
We are looking for exceptional early-career scientists conducting computational research with a proven record of accomplishment. The primary focus of this call is on candidates proposing research on b...
tinyurl.com
Reposted by Gräter lab
Reposted by Gräter lab
Reposted by Gräter lab
Gräter lab
@graeterlab.bsky.social
· Jan 28
Reposted by Gräter lab
Erik Poppleton
@floppleton.bsky.social
· Jan 17
Reposted by Gräter lab
Erik Poppleton
@floppleton.bsky.social
· Jan 27
Gräter lab
@graeterlab.bsky.social
· Jan 24

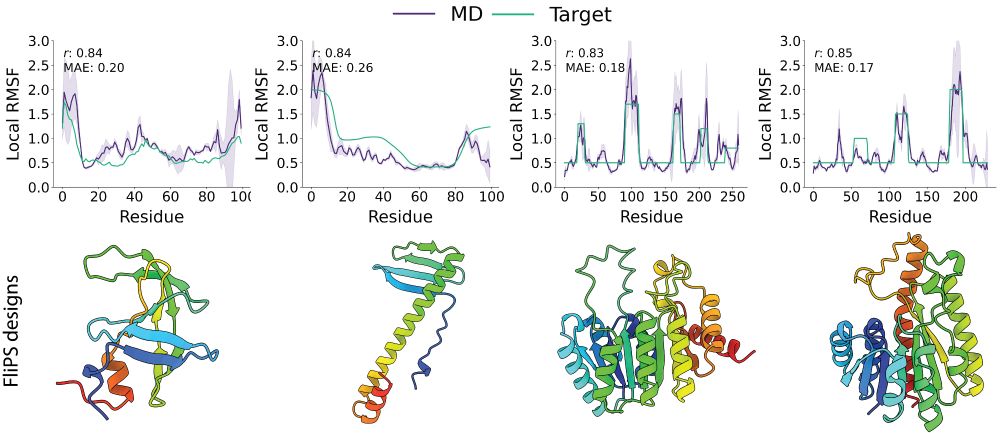
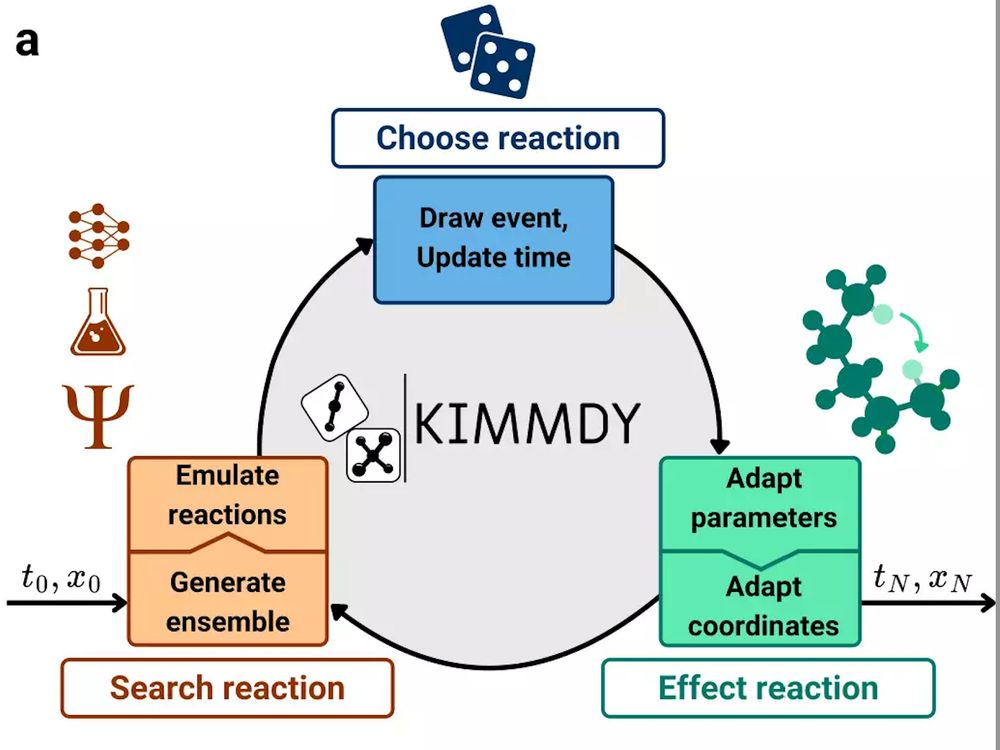
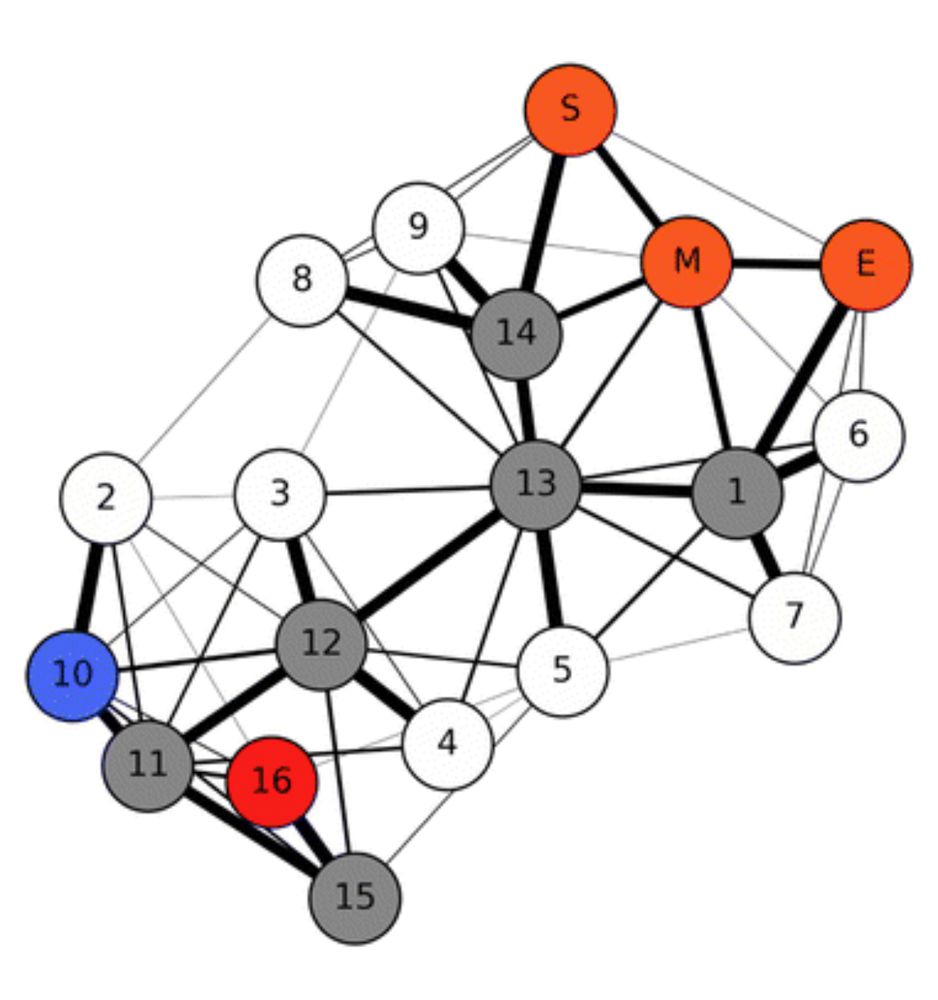
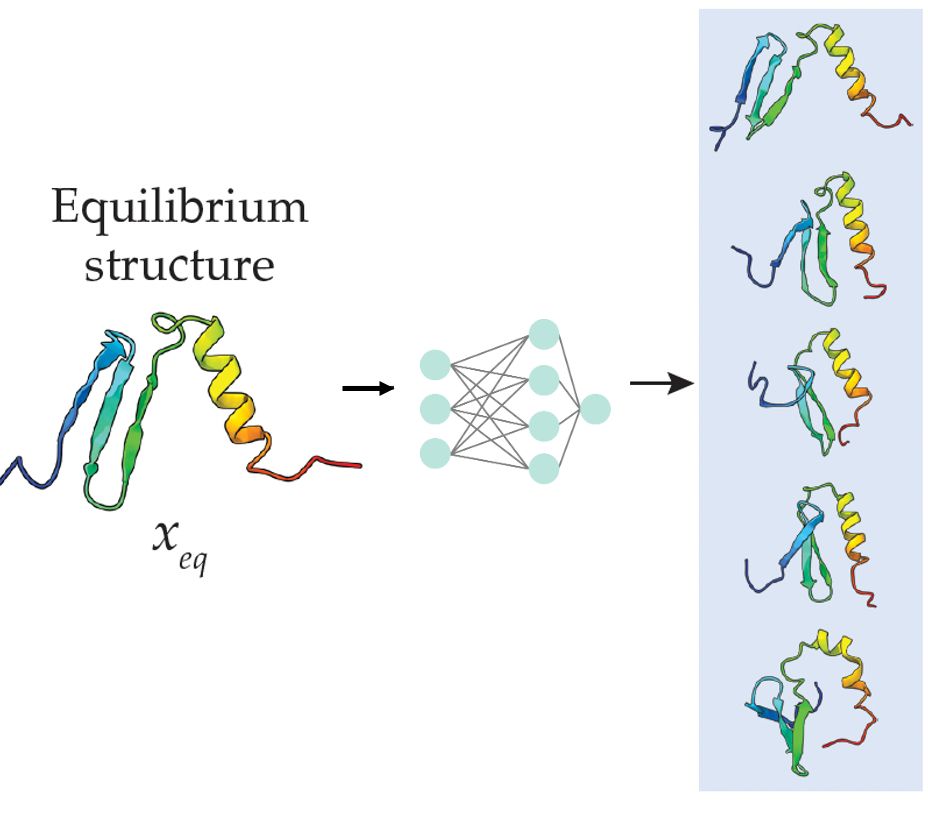
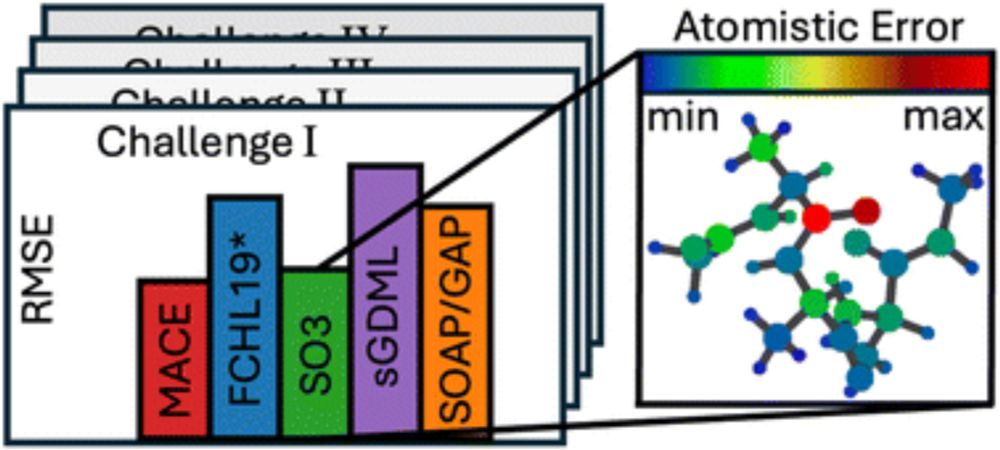
Grappa – a machine learned molecular mechanics force field
Simulating large molecular systems over long timescales requires force fields that are both accurate and efficient. In recent years, E(3) equivariant neural networks have lifted the tension between co...
pubs.rsc.org
Gräter lab
@graeterlab.bsky.social
· Jan 24

Grappa – a machine learned molecular mechanics force field
Simulating large molecular systems over long timescales requires force fields that are both accurate and efficient. In recent years, E(3) equivariant neural networks have lifted the tension between co...
pubs.rsc.org
Gräter lab
@graeterlab.bsky.social
· Jan 27

Gräter lab (@graeterlab.bsky.social)
We aim at unraveling the intricacies of molecular perception and designing sensing molecular systems from the ground up. We work at the interface of biophysics, biochemistry, and materials science.
@ Max Planck Institute for Polymer Research, Mainz.
bsky.app