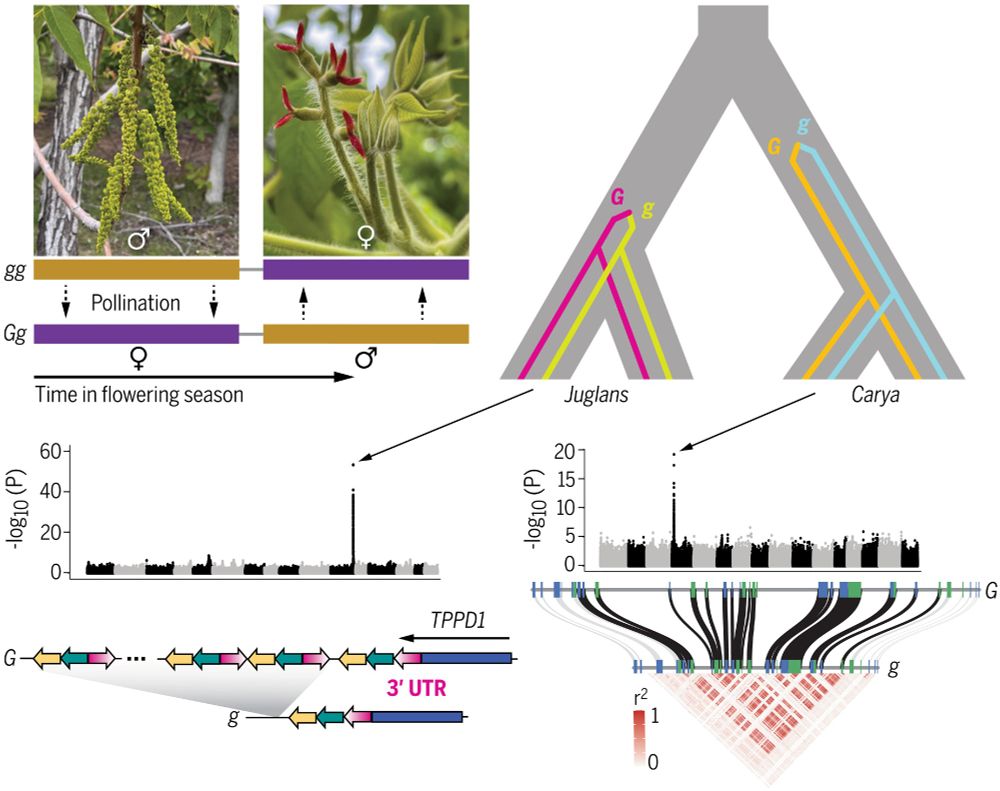
Greg Owens
@gregowens.bsky.social
1.8K followers
1.6K following
39 posts
Assistant Professor of Biology at the University of Victoria. Plant genomics and plant genomics jokes
Posts
Media
Videos
Starter Packs
Greg Owens
@gregowens.bsky.social
· Sep 1
Reposted by Greg Owens
Tyler Kent
@tylervkent.bsky.social
· Apr 7

Demographic history, genetic load, and the efficacy of selection in the globally invasive mosquito Aedes aegypti
Abstract. Aedes aegypti is the main vector species of yellow fever, dengue, Zika and chikungunya. The species is originally from Africa but has experienced
doi.org
Reposted by Greg Owens
Reposted by Greg Owens
Reposted by Greg Owens
Greg Owens
@gregowens.bsky.social
· Feb 11
Reposted by Greg Owens
Greg Owens
@gregowens.bsky.social
· Jan 19
hannah
@hannza.bsky.social
· Jan 19
Greg Owens
@gregowens.bsky.social
· Jan 17
Greg Owens
@gregowens.bsky.social
· Jan 17
Greg Owens
@gregowens.bsky.social
· Jan 17
Reposted by Greg Owens
Reposted by Greg Owens
Per Ahlberg
@perahlberg.bsky.social
· Dec 23
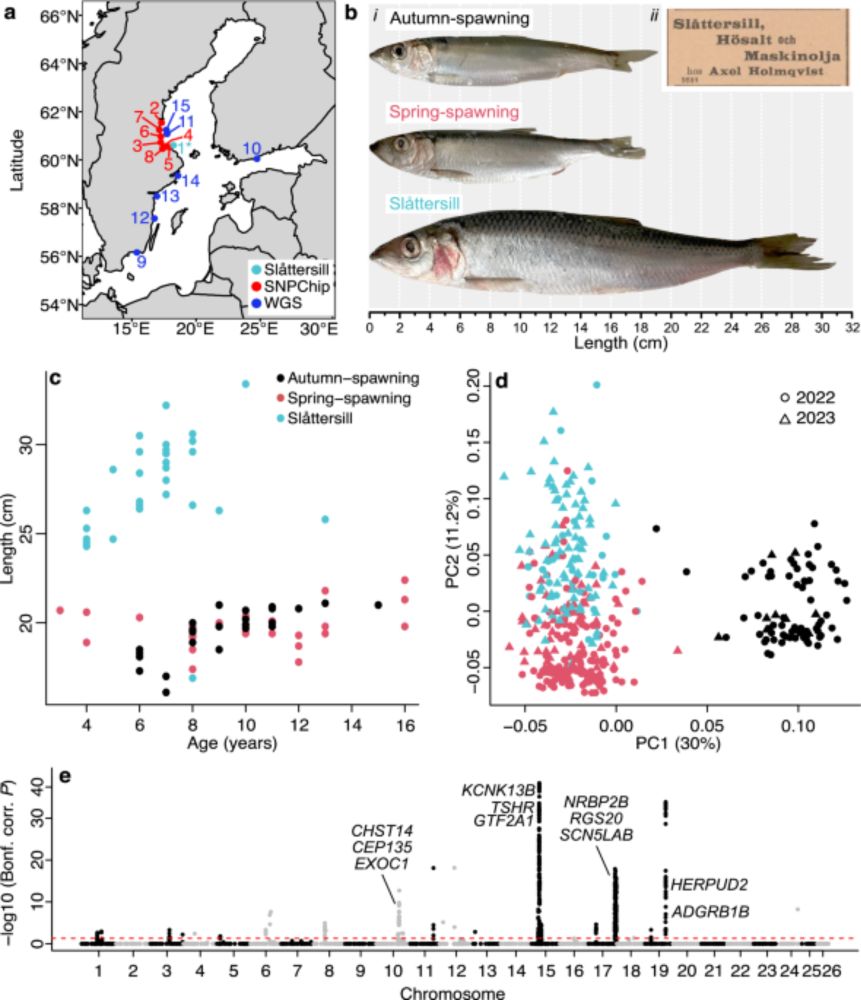
Evolution of fast-growing piscivorous herring in the young Baltic Sea - Nature Communications
The Atlantic herring is one of the world’s most abundant vertebrates and a typical plankton feeder of major ecological importance. This study shows that a piscivorous (fish-eating) ecotype of herring ...
www.nature.com
Reposted by Greg Owens
Reposted by Greg Owens
Jeff Spence
@jeffspence.github.io
· Dec 17

Specificity, length, and luck: How genes are prioritized by rare and common variant association studies
Standard genome-wide association studies (GWAS) and rare variant burden tests are essential tools for identifying trait-relevant genes. Although these methods are conceptually similar, we show by anal...
www.biorxiv.org
Reposted by Greg Owens
Ostevik Lab
@osteviklab.bsky.social
· Dec 7

Inversions contribute disproportionately to parallel genomic divergence in dune sunflowers - Nature Ecology & Evolution
Analysis of habitat data, quantitative trait locus mapping of seed size and selective sweeps show parallel selection acting on inversions in two independent dune ecotypes of the prairie sunflower, Hel...
www.nature.com