Helena Schulz-Mirbach
@helenasm.bsky.social
66 followers
68 following
16 posts
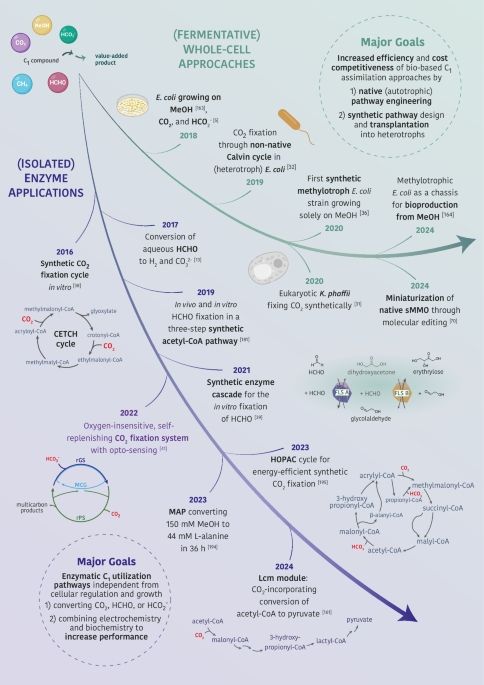
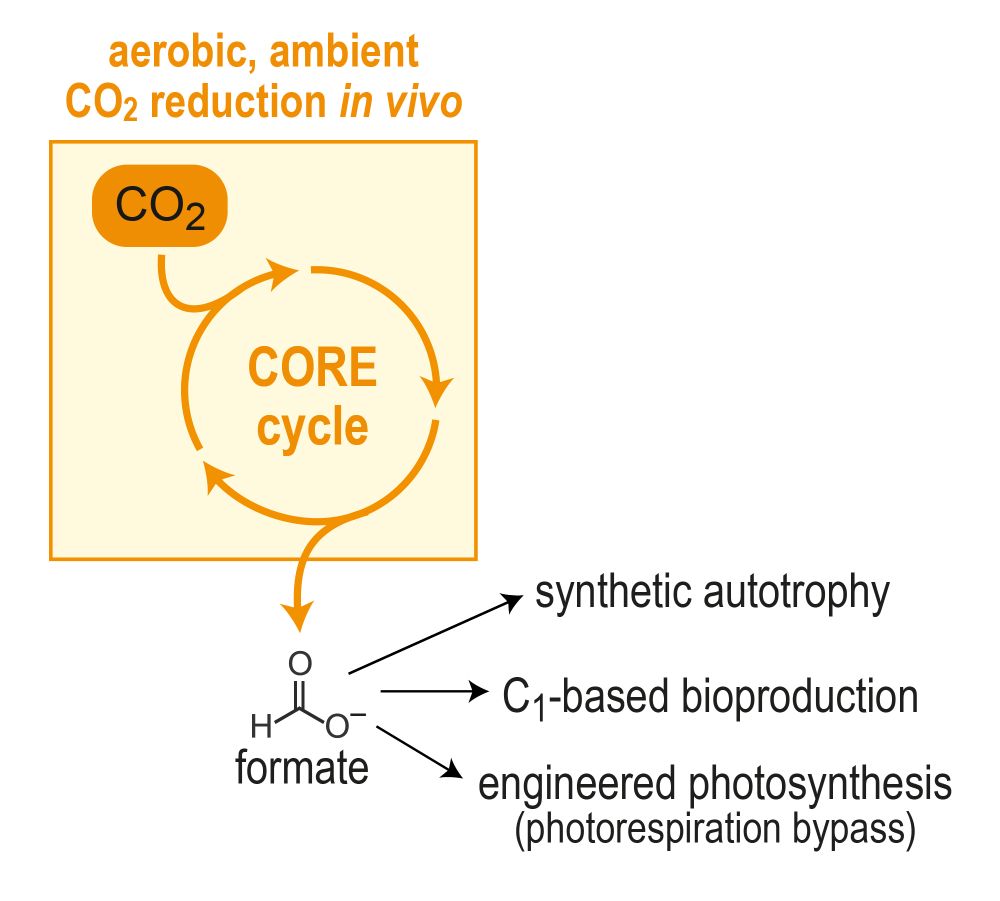
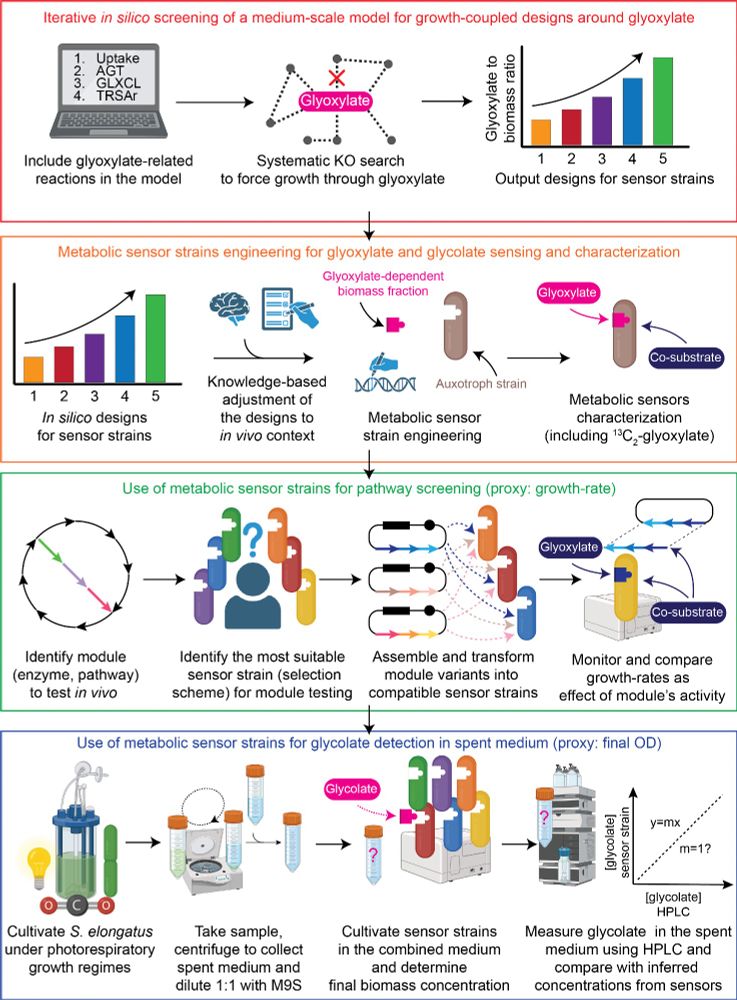
PhD student rewiring E. coli for synthetic C1 assimilation
Posts
Media
Videos
Starter Packs
Reposted by Helena Schulz-Mirbach
Reposted by Helena Schulz-Mirbach
Reposted by Helena Schulz-Mirbach
Reposted by Helena Schulz-Mirbach