www.nature.com/articles/s41...
www.nature.com/articles/s41...
Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/

Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/
Paper: doi.org/10.1101/2025...
Code: github.com/morrislab/mR...
A 🧵

Paper: doi.org/10.1101/2025...
Code: github.com/morrislab/mR...
A 🧵

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...



I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
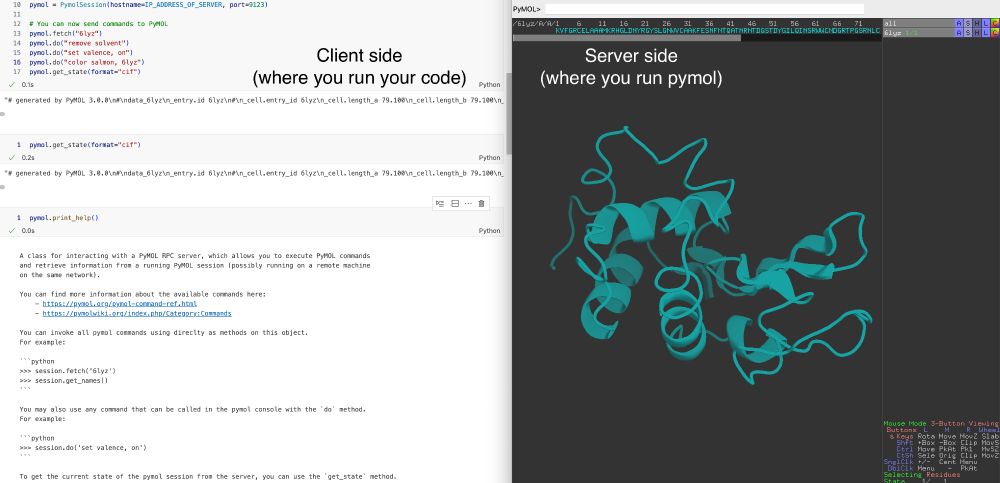
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`


