Hyejung Won
@hyejungwon.bsky.social
220 followers
71 following
24 posts
Associate Professor at UNC Chapel Hill
Neurogenetics
wonlab.org
Posts
Media
Videos
Starter Packs
Pinned
Hyejung Won
@hyejungwon.bsky.social
· Jan 27
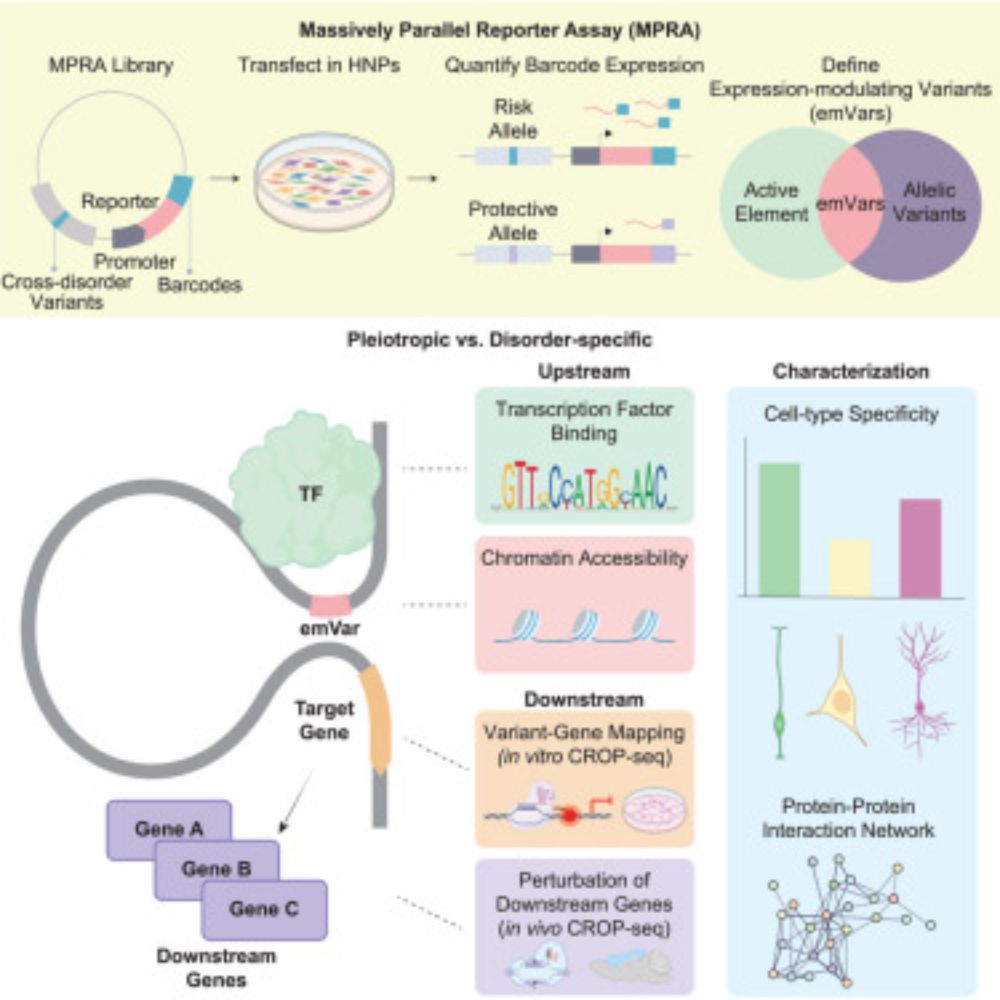
Massively parallel reporter assay investigates shared genetic variants of eight psychiatric disorders
High-throughput experimental validation of genetic variants linked to eight psychiatric
disorders reveals the regulatory mechanisms underlying variants with pleiotropic and
disorder-specific effects.
www.cell.com
Hyejung Won
@hyejungwon.bsky.social
· Jul 2
Hyejung Won
@hyejungwon.bsky.social
· Jul 2
Reposted by Hyejung Won
Reposted by Hyejung Won
Doug Fowler
@dougfowler.bsky.social
· May 15
Atlas of Variant Effects 2030 Roadmap: resolving human variants of uncertain significance
At the Clinical Atlas of Variant Effects meeting (CLAVE meeting, July 2024, Pittsburgh USA), we developed recommendations for a draft atlas that can be realized by 2030, with a focus on empowering gen...
zenodo.org
Reposted by Hyejung Won
Hyejung Won
@hyejungwon.bsky.social
· May 14
Hyejung Won
@hyejungwon.bsky.social
· May 1
Reposted by Hyejung Won
Martin Kampmann
@kampmann.bsky.social
· Apr 16
Takeshi Uenaka
@tuenaka.bsky.social
· Apr 15

Prevention of Transgene Silencing During Human Pluripotent Stem Cell Differentiation
While high and stable transgene expression can be achieved in undifferentiated pluripotent stem cells, conventional transgene expression systems are often silenced upon differentiation. Silencing occu...
www.biorxiv.org
Reposted by Hyejung Won
Hyejung Won
@hyejungwon.bsky.social
· Mar 6
Hyejung Won
@hyejungwon.bsky.social
· Feb 28

Genome-wide association study of borderline personality disorder reveals genetic overlap with bipolar disorder, major depression and schizophrenia - Translational Psychiatry
Translational Psychiatry - Genome-wide association study of borderline personality disorder reveals genetic overlap with bipolar disorder, major depression and schizophrenia
www.nature.com
Hyejung Won
@hyejungwon.bsky.social
· Feb 27
Reposted by Hyejung Won
Cedric Boeckx
@cedricboeckx.bsky.social
· Feb 12

Massively parallel assessment of gene regulatory activity at human cortical structure associated variants
Genetic association studies have identified hundreds of largely non-coding loci associated with inter-individual differences in the structure of the human cortex, though the specific genetic variants ...
www.biorxiv.org
Hyejung Won
@hyejungwon.bsky.social
· Jan 28
Hyejung Won
@hyejungwon.bsky.social
· Jan 28
Hyejung Won
@hyejungwon.bsky.social
· Jan 28
Hyejung Won
@hyejungwon.bsky.social
· Jan 28
Hyejung Won
@hyejungwon.bsky.social
· Jan 28
Hyejung Won
@hyejungwon.bsky.social
· Jan 28
Hyejung Won
@hyejungwon.bsky.social
· Jan 28
Hyejung Won
@hyejungwon.bsky.social
· Jan 27



