Jay Brito Querido
@jbquerido.bsky.social
3.4K followers
1.4K following
14 posts
Assistant Professor at the University of Michigan Medical School and @umlifesciences.bsky.social
#cryoEM #RNA #Ribosomes #RNAsky #RNABiology #Science
https://www.lsi.umich.edu/science/our-labs/brito-querido-lab
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Jay Brito Querido
Jay Brito Querido
@jbquerido.bsky.social
· Jul 23
Jay Brito Querido
@jbquerido.bsky.social
· Jul 23
Jay Brito Querido
@jbquerido.bsky.social
· Jul 23
Jay Brito Querido
@jbquerido.bsky.social
· Jul 22

SPIDR enables multiplexed mapping of RNA-protein interactions and uncovers a mechanism for selective translational suppression upon cell stress
SPIDR, a massively multiplexed method that simultaneously maps dozens of RNA-binding
proteins to their RNA targets at single-nucleotide resolution, uncovers new RNA-protein
interactions and provides c...
www.cell.com
Reposted by Jay Brito Querido
Chris H. Hill
@chillzaa.bsky.social
· Jul 19

A new protein-dependent riboswitch activates ribosomal frameshifting
Programmed -1 ribosomal frameshifting (PRF) is a ubiquitous translational control mechanism in RNA viruses, allowing them to change the relative abundance of proteins encoded in different reading fram...
www.biorxiv.org
Reposted by Jay Brito Querido
Alexey Amunts
@amunts.bsky.social
· Jul 14
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Kelly Nguyen
@kellythd-nguyen.bsky.social
· Jul 10
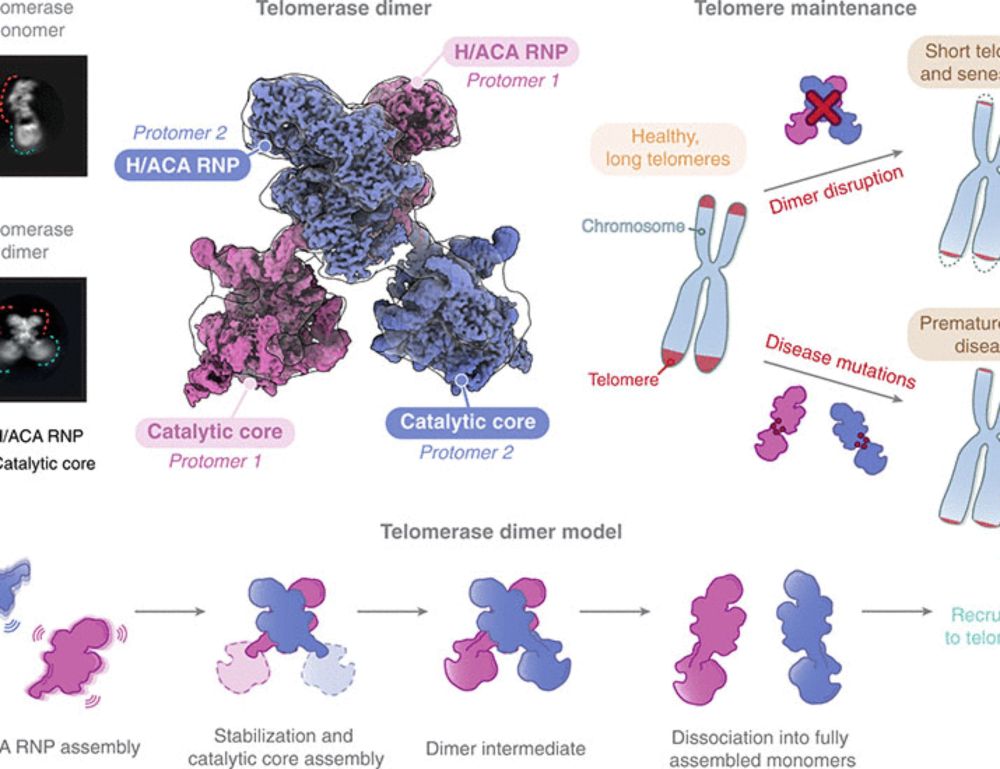
Cryo-EM structure of human telomerase dimer reveals H/ACA RNP-mediated dimerization
Telomerase ribonucleoprotein (RNP) synthesizes telomeric repeats at chromosome ends using a telomerase reverse transcriptase (TERT) and a telomerase RNA (hTR in humans). Previous structural work showe...
www.science.org
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Chelsey Spriggs
@drsprggs.bsky.social
· Jun 19
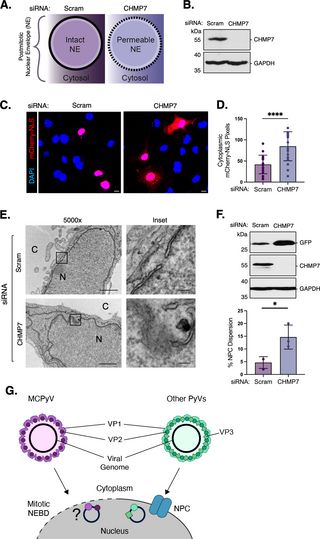
Temporal gating of nuclear import: How Merkel cell polyomavirus exploits the cell cycle for nuclear entry
Author summary Merkel cell polyomavirus (MCPyV) causes a rare but deadly form of skin cancer called Merkel cell carcinoma. However, many aspects of its infectious life cycle are unclear, including how...
journals.plos.org
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Merve Yigit
@merveyigit.bsky.social
· May 26
Sendoel Lab
@sendoellab.bsky.social
· May 26

The alternative initiation factor eIF2A regulates 40S subunit turnover in ribosome-associated quality control
The noncanonical translation initiation factor eIF2A plays critical roles in diverse cellular processes, including the integrated stress response, neurodegeneration and tumorigenesis. However, the pre...
www.biorxiv.org
Reposted by Jay Brito Querido
Sendoel Lab
@sendoellab.bsky.social
· May 26

The alternative initiation factor eIF2A regulates 40S subunit turnover in ribosome-associated quality control
The noncanonical translation initiation factor eIF2A plays critical roles in diverse cellular processes, including the integrated stress response, neurodegeneration and tumorigenesis. However, the pre...
www.biorxiv.org
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Yi-Wei Chang
@yiweichang.bsky.social
· May 15
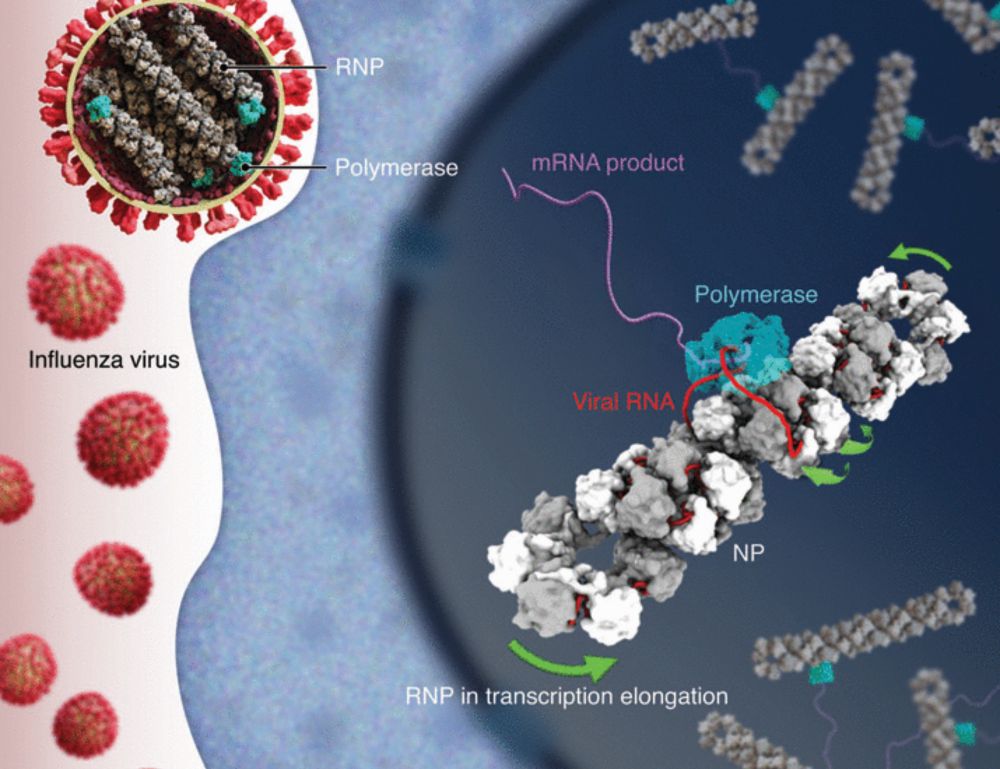
Molecular basis of influenza ribonucleoprotein complex assembly and processive RNA synthesis
Influenza viruses replicate and transcribe their genome in the context of a conserved ribonucleoprotein (RNP) complex. By integrating cryo–electron microscopy single-particle analysis and cryo–electro...
www.science.org
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Reposted by Jay Brito Querido
Wilson Lab
@wilsonlab.bsky.social
· May 7

The structure of the Vibrio natriegens 70S ribosome in complex with the proline-rich antimicrobial peptide Bac5(1–17)
Abstract. Proline-rich antimicrobial peptides (PrAMPs) are produced as part of the innate immune response of animals, insects, and plants. The well-charact
academic.oup.com
Reposted by Jay Brito Querido
Alan Brown
@alanbrownhms.bsky.social
· Apr 30
Sven Lange
@sven-m-lange.bsky.social
· Apr 29

A conserved mechanism for the retrieval of polyubiquitinated proteins from cilia
The temporospatial distribution of proteins within cilia is regulated by intraflagellar transport (IFT), wherein molecular trains shuttle between the cell body and cilium. Defects in this process impair various signal-transduction pathways and cause ciliopathies. Although K63-linked ubiquitination appears to trigger protein export from cilia, the mechanisms coupling polyubiquitinated proteins to IFT remain unclear. Using a multidisciplinary approach, we demonstrate that a complex of CFAP36, a conserved ciliary protein of previously unknown function, and ARL3, a GTPase involved in ciliary import, binds polyubiquitinated proteins and links them to retrograde IFT trains. CFAP36 uses a coincidence detection mechanism to simultaneously bind two IFT subunits accessible only in retrograde trains. Depleting CFAP36 accumulates K63-linked ubiquitin in cilia and disrupts Hedgehog signaling, a pathway reliant on the retrieval of ubiquitinated receptors. These findings advance our understanding of ubiquitin-mediated protein transport and ciliary homeostasis, and demonstrate how structural changes in IFT trains achieve cargo selectivity. ### Competing Interest Statement The authors have declared no competing interest. Sara Elizabeth O'Brien Trust Postdoctoral Fellowship awarded through the Charles A. King Trust Postdoctoral Research Fellowship Program, , 8460873-01 Richard and Susan Smith Family Foundation, https://ror.org/05j95n956, National Institute of General Medical Sciences (NIGMS), , R01GM141109, R01GM143183
www.biorxiv.org