Krzysztof Herka
@kherka.bsky.social
66 followers
180 following
16 posts
PhD student, Balasubramanian lab, Cancer Research UK - Cambridge Institute (@cruk-ci.bsky.social)
Posts
Media
Videos
Starter Packs
Pinned
Krzysztof Herka
@kherka.bsky.social
· Jun 4
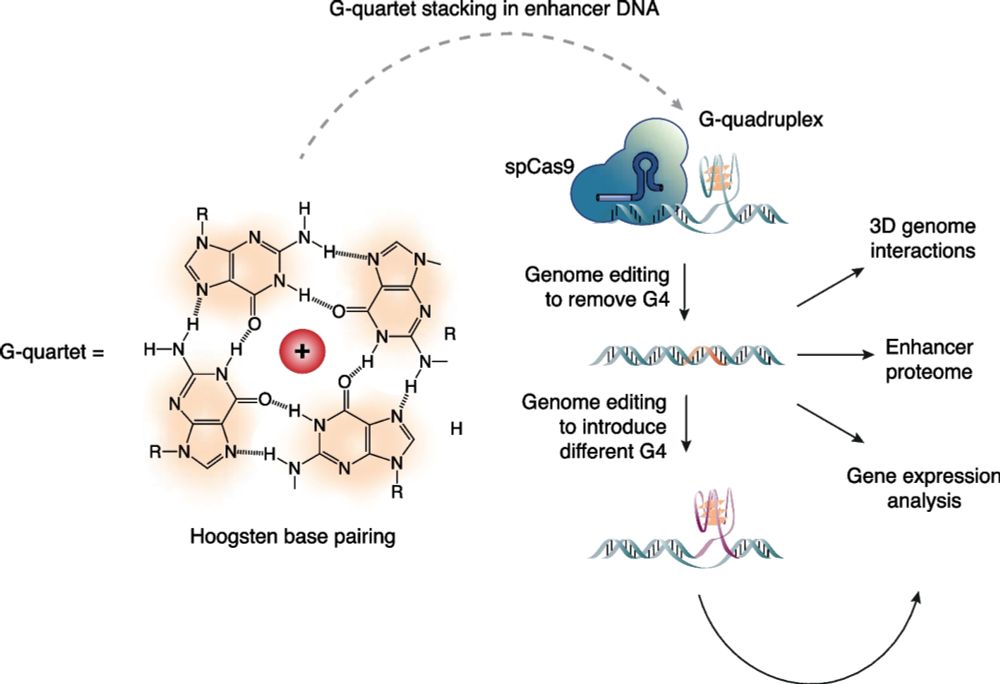
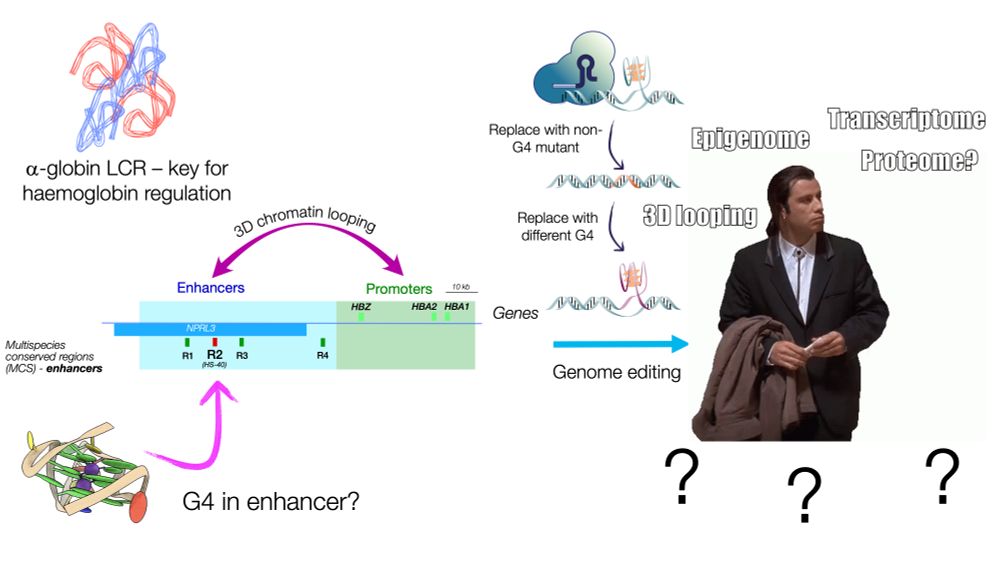
DNA G-quadruplex structures act as functional elements in α- and β-globin enhancers - Genome Biology
Background Enhancer elements interact with target genes at a distance to modulate their expression, but the molecular details of enhancer–promoter interaction are incompletely understood. G-quadruplex...
doi.org
Krzysztof Herka
@kherka.bsky.social
· Sep 2
Sean Flynn
@seanflynn.bsky.social
· Sep 2
Architects of the Genome: Using genomic technologies to elucidate the role of CTCF in neurodevelopmental disorders - GW4 BioMed MRC DTP
Project Code MRCNMH26Ex Flynn Project Type The project is primarily wet-lab based, with a dry-lab component in Years 2 and 3. Research Theme Neuroscience and Mental Health Project Summary Download Sum...
gw4biomed.ac.uk
Reposted by Krzysztof Herka
Krzysztof Herka
@kherka.bsky.social
· Jun 7
Krzysztof Herka
@kherka.bsky.social
· Jun 4
Krzysztof Herka
@kherka.bsky.social
· Jun 4
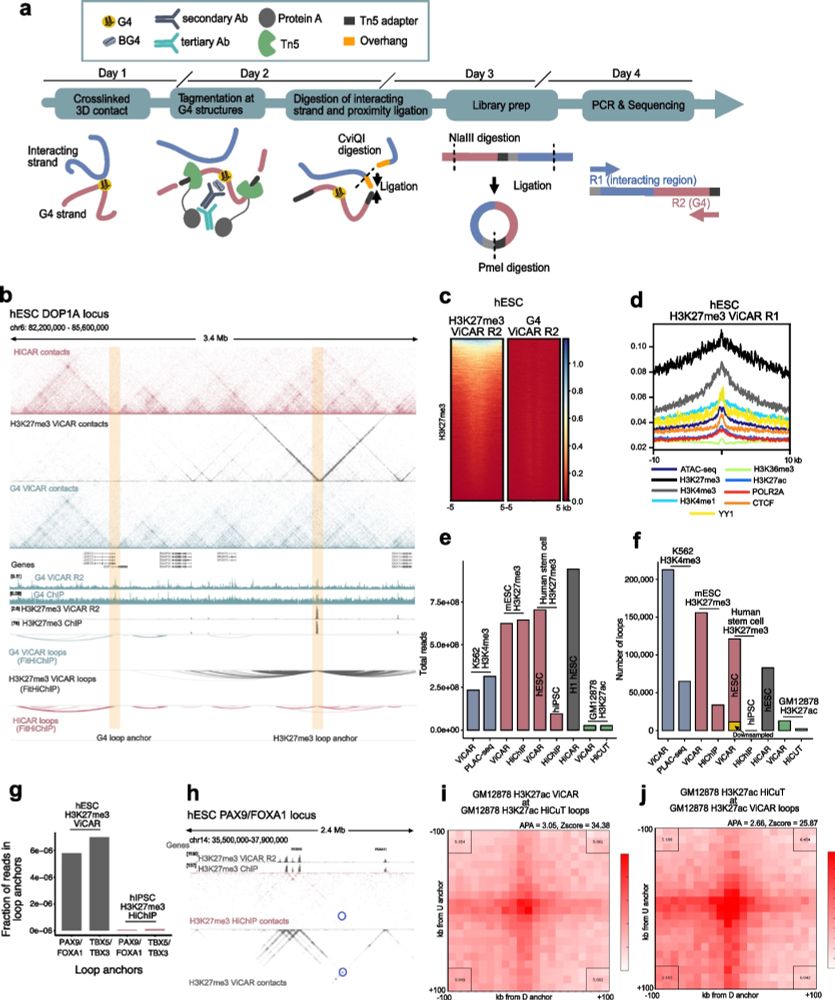
Improved simultaneous mapping of epigenetic features and 3D chromatin structure via ViCAR - Genome Biology
Methods to measure chromatin contacts at genomic regions bound by histone modifications or proteins are important tools to investigate chromatin organization. However, such methods do not capture the ...
genomebiology.biomedcentral.com
Krzysztof Herka
@kherka.bsky.social
· Jun 4
Krzysztof Herka
@kherka.bsky.social
· Jun 4
Krzysztof Herka
@kherka.bsky.social
· Jun 4
Krzysztof Herka
@kherka.bsky.social
· Jun 4

DNA G-quadruplex structures act as functional elements in α- and β-globin enhancers - Genome Biology
Background Enhancer elements interact with target genes at a distance to modulate their expression, but the molecular details of enhancer–promoter interaction are incompletely understood. G-quadruplex...
doi.org
Reposted by Krzysztof Herka
Reposted by Krzysztof Herka
Reposted by Krzysztof Herka
Krzysztof Herka
@kherka.bsky.social
· Nov 15
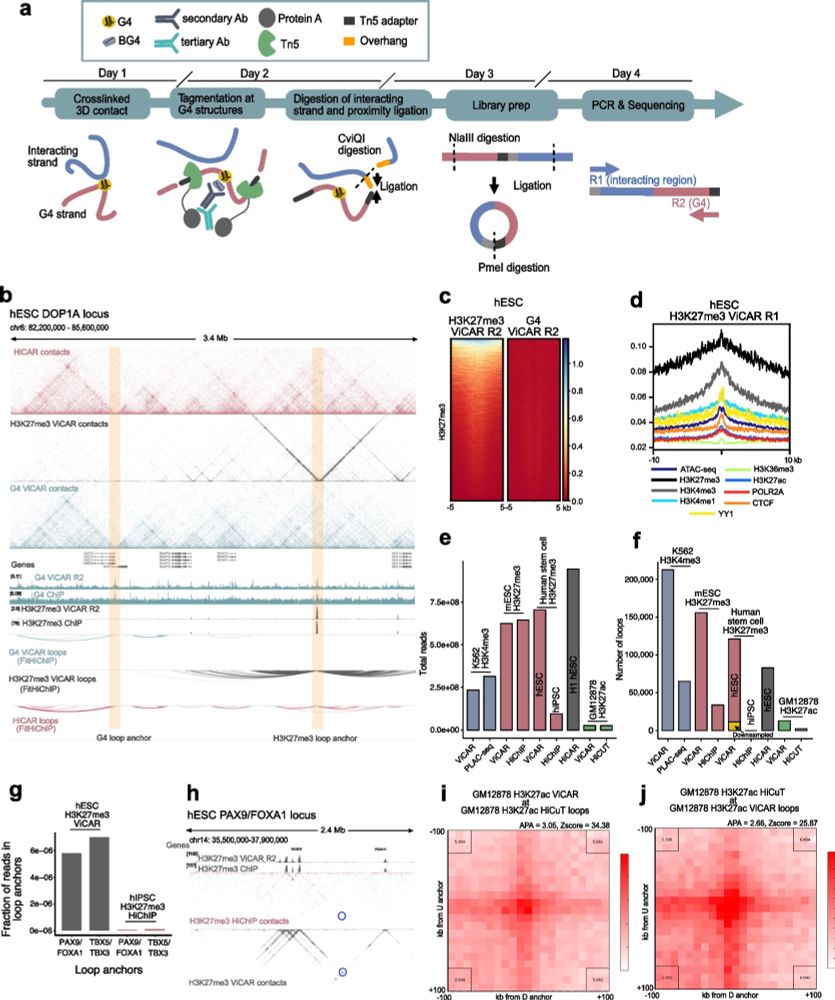
Improved simultaneous mapping of epigenetic features and 3D chromatin structure via ViCAR - Genome Biology
Methods to measure chromatin contacts at genomic regions bound by histone modifications or proteins are important tools to investigate chromatin organization. However, such methods do not capture the ...
genomebiology.biomedcentral.com
Krzysztof Herka
@kherka.bsky.social
· Nov 15