Koushik
@koushikn.bsky.social
Data Scientist Generative AI @BayerCropScience. ML for Plant Biology. PhD @IowaStateUniversity https://www.linkedin.com/in/koushik-nagasubramanian/
Reposted by Koushik
@jengreitz.bsky.social l & my lab want to co-hire a computational biologist/biostatistician with project management expertise to help map the regulatory code of the human genome and discover genetic mechanisms of disease.
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT


August 19, 2025 at 12:29 AM
@jengreitz.bsky.social l & my lab want to co-hire a computational biologist/biostatistician with project management expertise to help map the regulatory code of the human genome and discover genetic mechanisms of disease.
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
Reposted by Koushik
In 1965, Margaret Dayhoff published the Atlas of Protein Sequence and Structure, which collated the 65 proteins whose amino acid sequences were then known.
Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
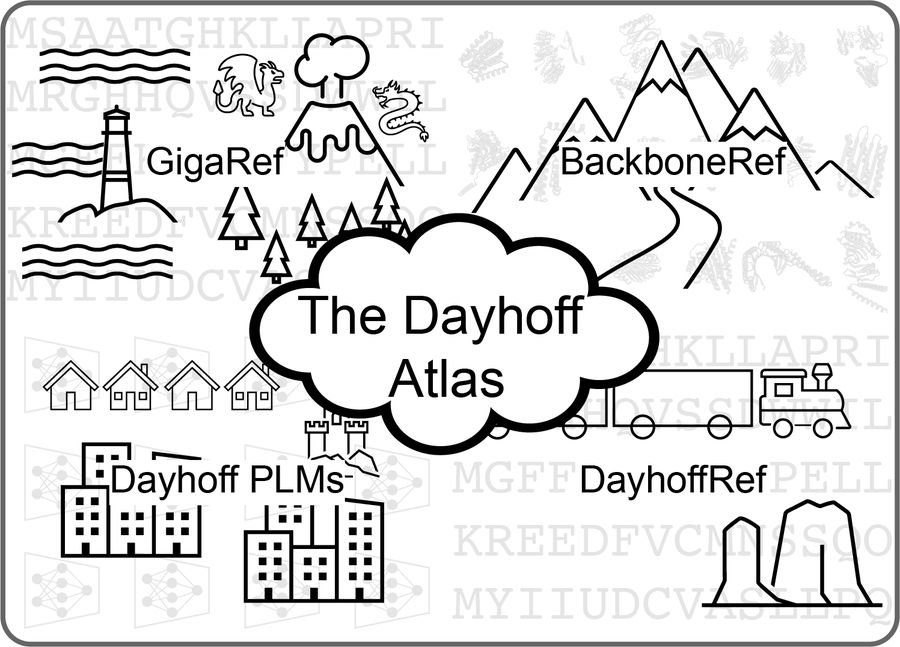
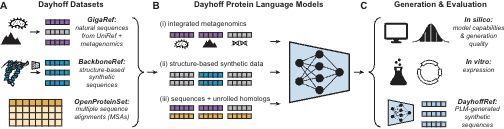
July 25, 2025 at 10:05 PM
In 1965, Margaret Dayhoff published the Atlas of Protein Sequence and Structure, which collated the 65 proteins whose amino acid sequences were then known.
Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
Reposted by Koushik
An assessment of DNA language models concludes:
◼️ They do not offer compelling gains over baseline models
Their performance is inconsistent and requires much more compute.
arxiv.org/abs/2412.05430
◼️ They do not offer compelling gains over baseline models
Their performance is inconsistent and requires much more compute.
arxiv.org/abs/2412.05430

June 23, 2025 at 8:21 PM
An assessment of DNA language models concludes:
◼️ They do not offer compelling gains over baseline models
Their performance is inconsistent and requires much more compute.
arxiv.org/abs/2412.05430
◼️ They do not offer compelling gains over baseline models
Their performance is inconsistent and requires much more compute.
arxiv.org/abs/2412.05430
Reposted by Koushik
Our structural core gene pipeline Unicode is now published at GBE
📄 doi.org/10.1093/gbe/...
Please also check out @dongwookkim.bsky.social’s
🧵 bsky.app/profile/dong...
📄 doi.org/10.1093/gbe/...
Please also check out @dongwookkim.bsky.social’s
🧵 bsky.app/profile/dong...
June 3, 2025 at 5:19 PM
Our structural core gene pipeline Unicode is now published at GBE
📄 doi.org/10.1093/gbe/...
Please also check out @dongwookkim.bsky.social’s
🧵 bsky.app/profile/dong...
📄 doi.org/10.1093/gbe/...
Please also check out @dongwookkim.bsky.social’s
🧵 bsky.app/profile/dong...
Reposted by Koushik
The only reason you love chocolate is because of FUNGUS.
Cacao seeds contain high amounts of polyphenols, making them intensely bitter & unpleasant. There are two natural fungi that do the heavy lifting in turning them into chocolate.
Let's do a quick tour of the process of chocolate making.
Cacao seeds contain high amounts of polyphenols, making them intensely bitter & unpleasant. There are two natural fungi that do the heavy lifting in turning them into chocolate.
Let's do a quick tour of the process of chocolate making.

May 26, 2025 at 9:19 PM
The only reason you love chocolate is because of FUNGUS.
Cacao seeds contain high amounts of polyphenols, making them intensely bitter & unpleasant. There are two natural fungi that do the heavy lifting in turning them into chocolate.
Let's do a quick tour of the process of chocolate making.
Cacao seeds contain high amounts of polyphenols, making them intensely bitter & unpleasant. There are two natural fungi that do the heavy lifting in turning them into chocolate.
Let's do a quick tour of the process of chocolate making.
Reposted by Koushik
Three BioML starter packs now!
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
December 3, 2024 at 3:27 AM
Three BioML starter packs now!
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
Reposted by Koushik
AFESM: a metagenomic guide through the protein structure universe! We clustered 821M structures (AFDB&ESMatlas) into 5.12M groups; revealing biome-specific groups, only 1 new fold even after AlphaFold2 re-prediction & many novel domain combos. 🧵
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

April 27, 2025 at 12:13 AM
AFESM: a metagenomic guide through the protein structure universe! We clustered 821M structures (AFDB&ESMatlas) into 5.12M groups; revealing biome-specific groups, only 1 new fold even after AlphaFold2 re-prediction & many novel domain combos. 🧵
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
Reposted by Koushik
Super excited to share our review on genomic deep learning models for non-coding variant effect prediction, with Ayesha Bajwa and Nilah Ioannidis. We’d like this review to be a useful resource, and welcome any feedback, comments, or questions! 1/4
arxiv.org/abs/2411.11158
arxiv.org/abs/2411.11158

Leveraging genomic deep learning models for non-coding variant effect prediction
The majority of genetic variants identified in genome-wide association studies of complex traits are non-coding, and characterizing their function remains an important challenge in human genetics. Gen...
arxiv.org
November 20, 2024 at 1:31 AM
Super excited to share our review on genomic deep learning models for non-coding variant effect prediction, with Ayesha Bajwa and Nilah Ioannidis. We’d like this review to be a useful resource, and welcome any feedback, comments, or questions! 1/4
arxiv.org/abs/2411.11158
arxiv.org/abs/2411.11158
Reposted by Koushik
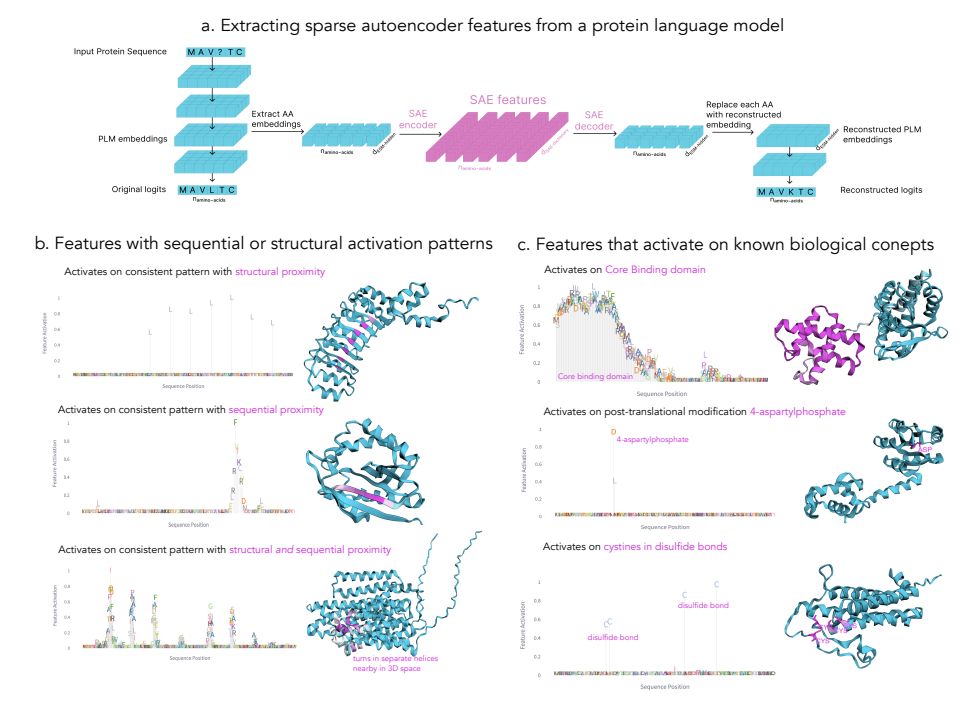
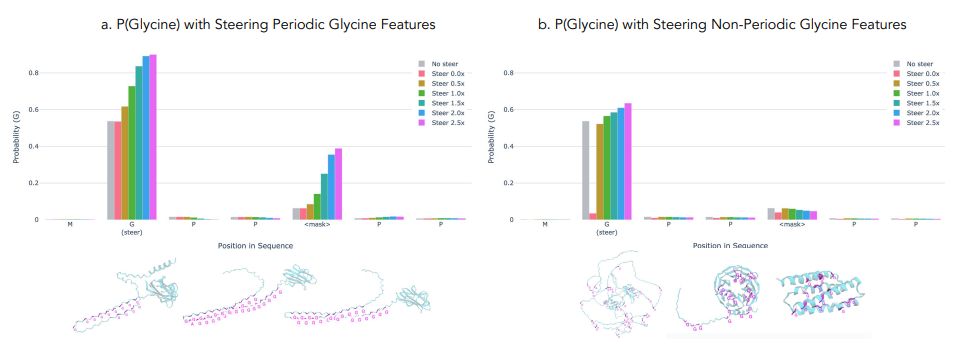
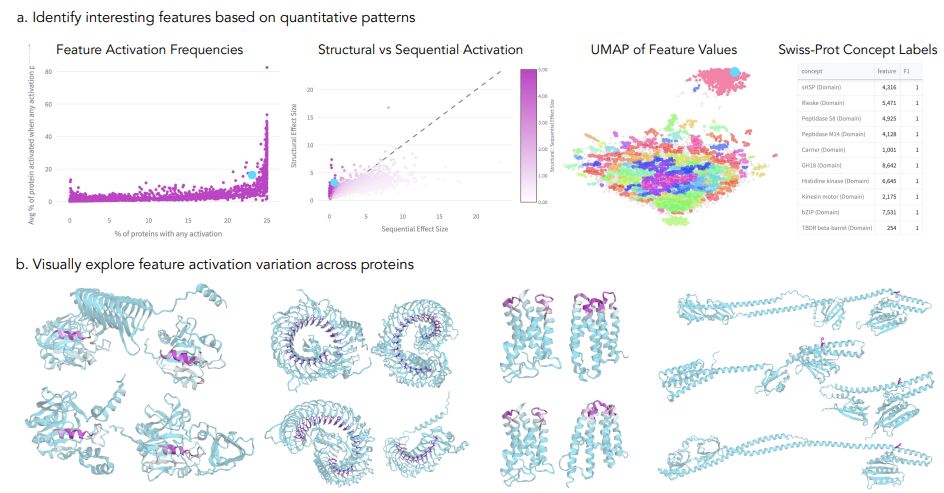
November 18, 2024 at 10:20 PM
Reposted by Koushik
Two BioML starter packs now:
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
Anybody have a bioml starter pack?
November 18, 2024 at 5:09 PM
Two BioML starter packs now:
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
Reposted by Koushik
DEGU distills an ensemble of models into a single model, retaining the ensemble’s predictive performance while providing uncertainty estimates - ie both epistemic (or model) and aleatoric (or data) uncertainty.
Led by @zrcjessica
Paper: www.biorxiv.org/content/10.1...
2/n
Led by @zrcjessica
Paper: www.biorxiv.org/content/10.1...
2/n

Uncertainty-aware genomic deep learning with knowledge distillation
Deep neural networks (DNNs) have advanced predictive modeling for regulatory genomics, but challenges remain in ensuring the reliability of their predictions and understanding the key factors behind t...
www.biorxiv.org
November 16, 2024 at 4:14 PM
DEGU distills an ensemble of models into a single model, retaining the ensemble’s predictive performance while providing uncertainty estimates - ie both epistemic (or model) and aleatoric (or data) uncertainty.
Led by @zrcjessica
Paper: www.biorxiv.org/content/10.1...
2/n
Led by @zrcjessica
Paper: www.biorxiv.org/content/10.1...
2/n
Reposted by Koushik
Large protein language models can learn complex epistatic interactions, but how much does that help with predicting variant effects? In this NeurIPS article, we show that classical independent-sites phylogenetic models can outperform pLMs on this task.
1/7
openreview.net/forum?id=H7m...
1/7
openreview.net/forum?id=H7m...

Ultrafast classical phylogenetic method beats large protein...
Amino acid substitution rate matrices are fundamental to statistical phylogenetics and evolutionary biology. Estimating them typically requires reconstructed trees for massive amounts of aligned...
openreview.net
November 16, 2024 at 8:42 PM
Large protein language models can learn complex epistatic interactions, but how much does that help with predicting variant effects? In this NeurIPS article, we show that classical independent-sites phylogenetic models can outperform pLMs on this task.
1/7
openreview.net/forum?id=H7m...
1/7
openreview.net/forum?id=H7m...
Reposted by Koushik
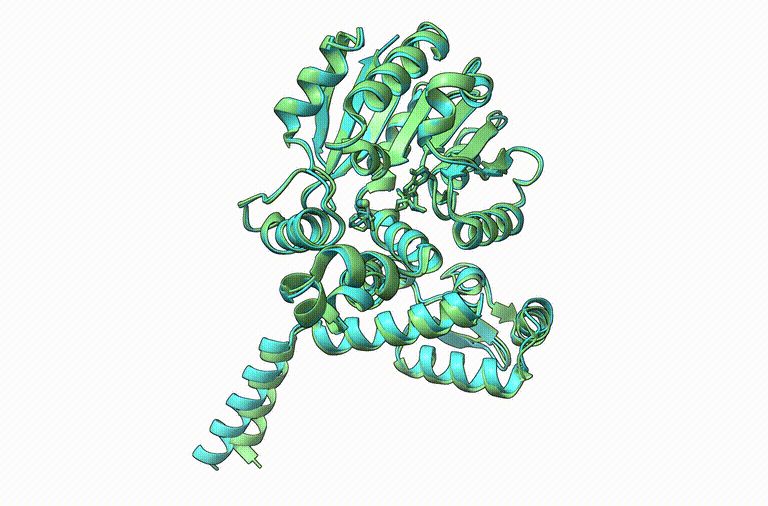
Thrilled to announce Boltz-1, the first open-source and commercially available model to achieve AlphaFold3-level accuracy on biomolecular structure prediction! An exciting collaboration with Jeremy, Saro, and an amazing team at MIT and Genesis Therapeutics. A thread!
November 17, 2024 at 4:20 PM
Thrilled to announce Boltz-1, the first open-source and commercially available model to achieve AlphaFold3-level accuracy on biomolecular structure prediction! An exciting collaboration with Jeremy, Saro, and an amazing team at MIT and Genesis Therapeutics. A thread!
Reposted by Koushik
Anybody have a bioml starter pack?
November 11, 2024 at 11:45 PM

