Leon Hilgers
@leonhilgers.bsky.social
250 followers
440 following
15 posts
Postdoc
@LOEWE_TBG @Senckenberg. Evolutionary genomics, gene expression, global change, novelties, climate, turtles & eels!
He/him
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Xueling (Ling) Yi
@xuelingyi.bsky.social
· Aug 17
Reposted by Leon Hilgers
Leon Hilgers
@leonhilgers.bsky.social
· Aug 2
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Dr. Carly Kenkel
@drcarl.bsky.social
· May 15
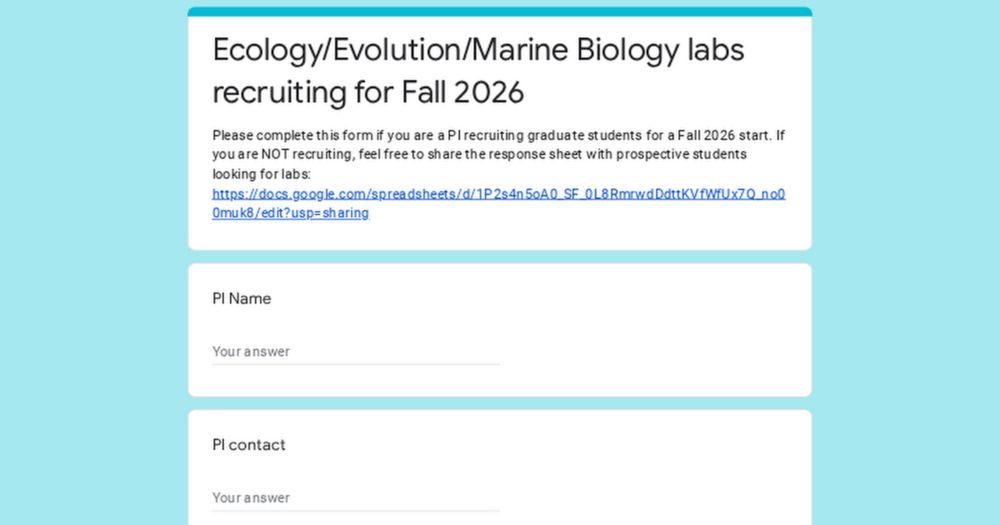
Ecology/Evolution/Marine Biology labs recruiting for Fall 2026
Please complete this form if you are a PI recruiting graduate students for a Fall 2026 start. If you are NOT recruiting, feel free to share the response sheet with prospective students looking for lab...
forms.gle
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Bernhard Bein
@bernhardbein.bsky.social
· Feb 10
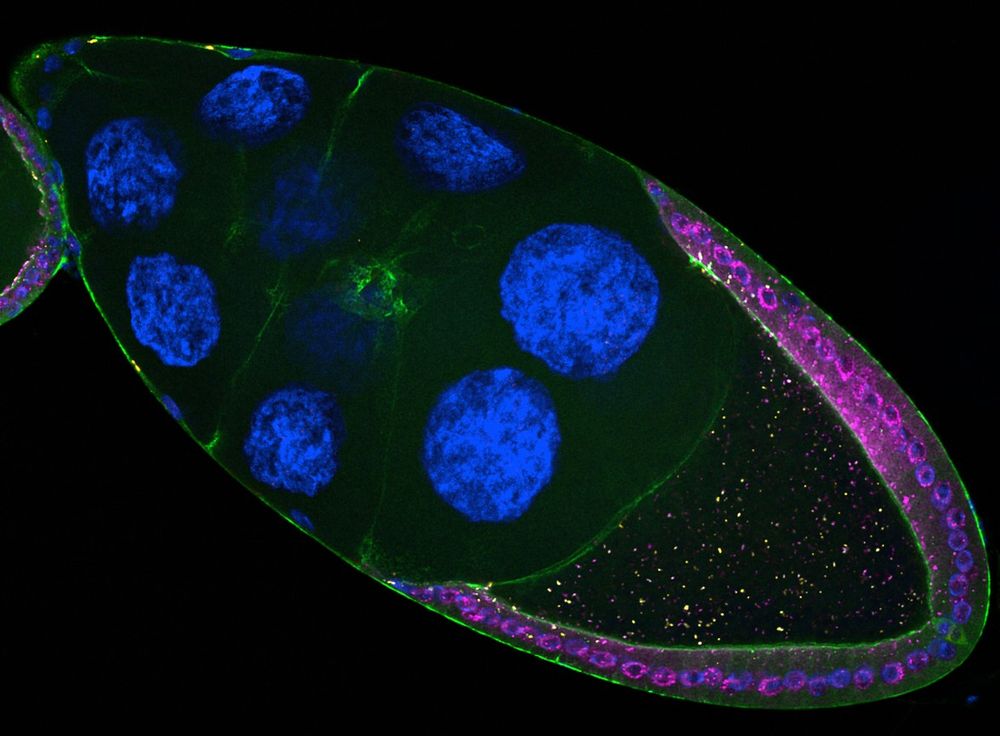
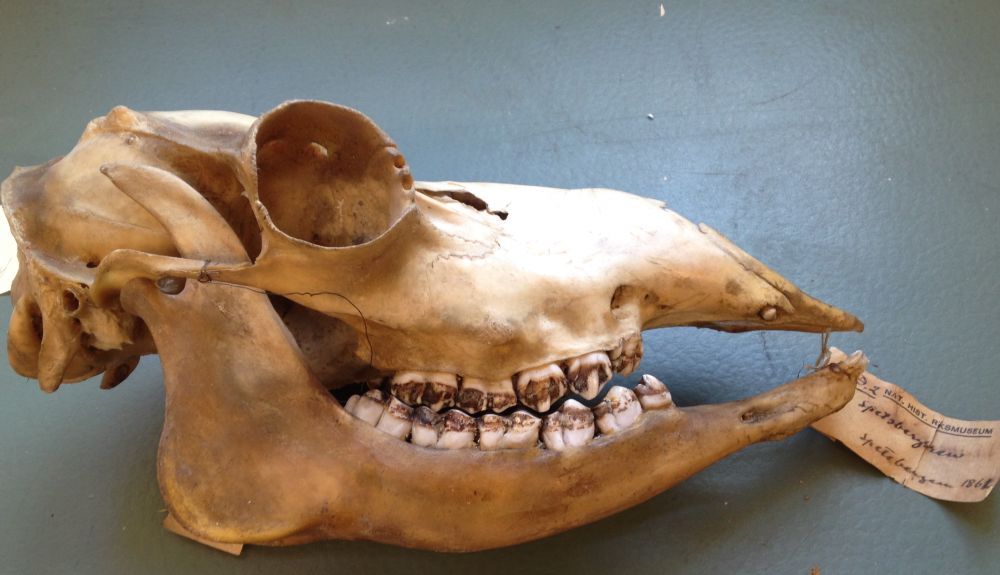
Long-read sequencing and genome assembly of natural history collection samples and challenging specimens - Genome Biology
Museum collections harbor millions of samples, largely unutilized for long-read sequencing. Here, we use ethanol-preserved samples containing kilobase-sized DNA to show that amplification-free protoco...
genomebiology.biomedcentral.com
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Reposted by Leon Hilgers
Leon Hilgers
@leonhilgers.bsky.social
· Feb 7