Bernhard Bein
@bernhardbein.bsky.social
29 followers
59 following
13 posts
PhD student in comparative genomics @ Senckenberg and Goethe University Frankfurt
Posts
Media
Videos
Starter Packs
Pinned
Bernhard Bein
@bernhardbein.bsky.social
· Feb 10
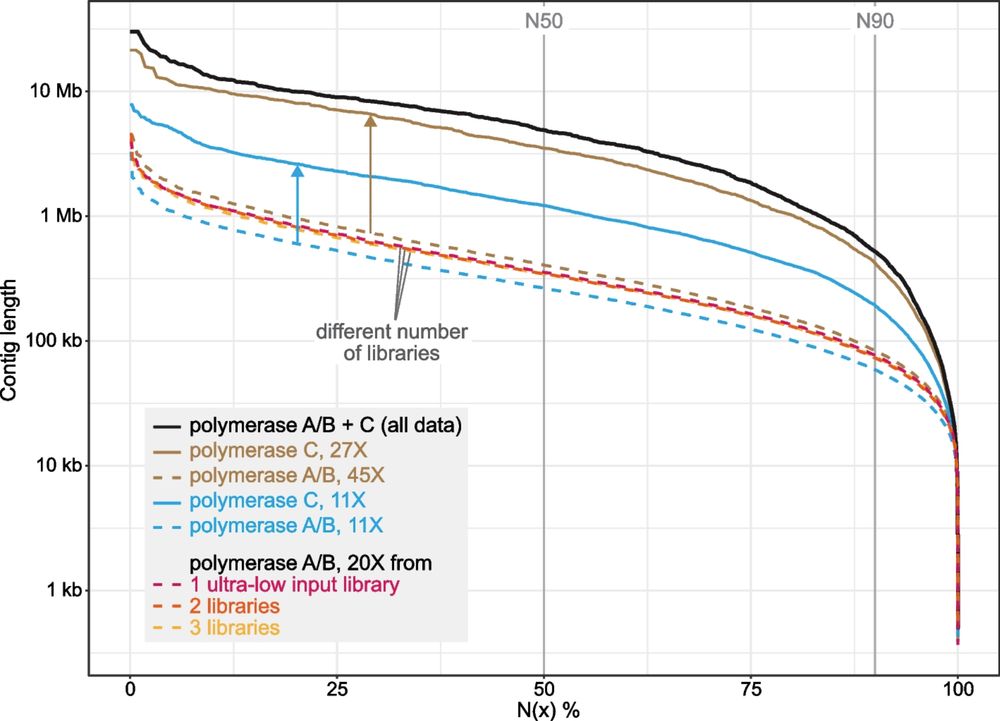
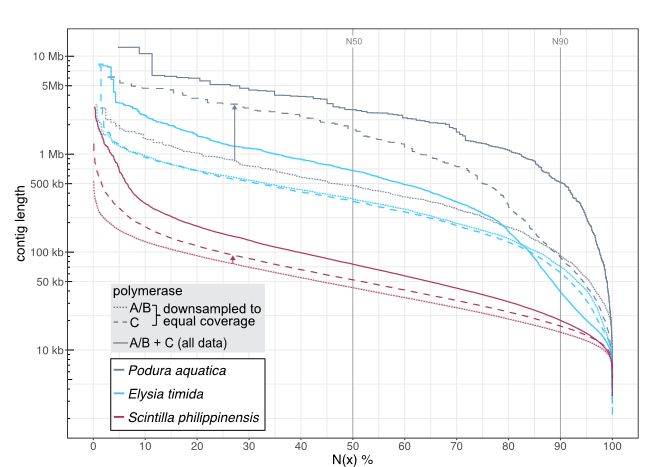
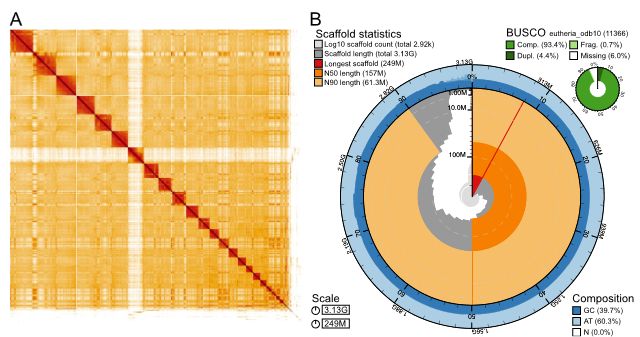
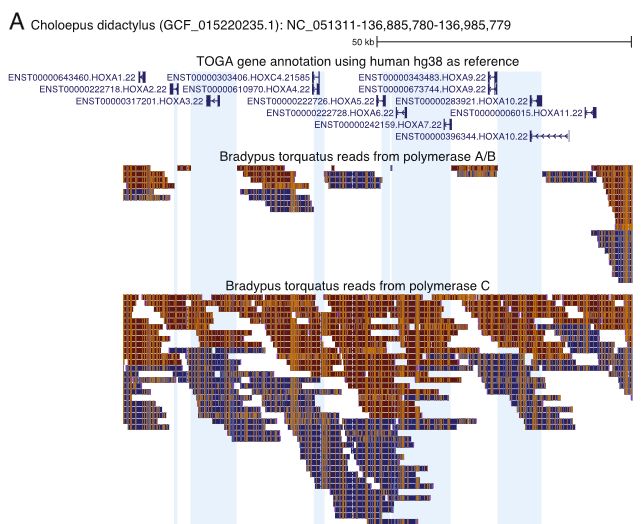
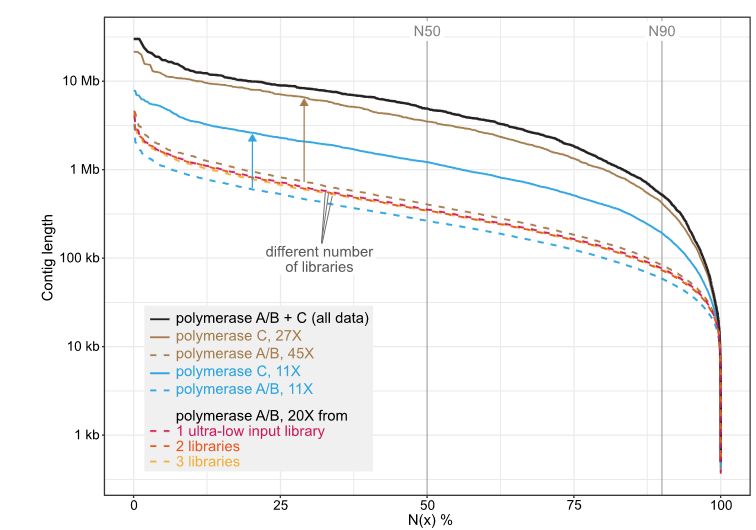
Long-read sequencing and genome assembly of natural history collection samples and challenging specimens - Genome Biology
Museum collections harbor millions of samples, largely unutilized for long-read sequencing. Here, we use ethanol-preserved samples containing kilobase-sized DNA to show that amplification-free protoco...
genomebiology.biomedcentral.com
Reposted by Bernhard Bein
Reposted by Bernhard Bein
Reposted by Bernhard Bein
mara lawniczak
@marakat.bsky.social
· Jul 7
Reposted by Bernhard Bein
Phil Morin
@philmor1964.bsky.social
· Jul 3

Frontiers | Genomic infrastructure for cetacean research and conservation: reference genomes for eight families spanning the cetacean tree of life
Reference genomes from representative species across families provide the critical infrastructure for research and conservation. The Cetacean Genomes Project...
www.frontiersin.org
Bernhard Bein
@bernhardbein.bsky.social
· May 20
PacBio
@pacbio.bsky.social
· May 19

Cancer research gains a powerful new tool as Ampli-Fi makes HiFi sequencing possible for FFPE-preserved samples - PacBio
Cancer research depends on the details we can actually see. And, until now, FFPE samples have not been compatible with long-read sequencing. That’s changing. This year at AACR, we...
bit.ly
Reposted by Bernhard Bein
Reposted by Bernhard Bein
Bernhard Bein
@bernhardbein.bsky.social
· Mar 21

Evaluating the Benefits and Limits of Multiple Displacement Amplification With Whole‐Genome Oxford Nanopore Sequencing
Multiple displacement amplification (MDA) outperforms conventional PCR in long fragment and whole-genome amplification, making it attractive to couple MDA with long-read sequencing of samples with li...
onlinelibrary.wiley.com
Reposted by Bernhard Bein
Bernhard Bein
@bernhardbein.bsky.social
· Feb 24
Reposted by Bernhard Bein
Bernhard Bein
@bernhardbein.bsky.social
· Feb 10