ORCID: 0009-0006-5724-8121
github: https://github.com/hanjinliu
We show that human spastin, a microtubule severing enzyme, behaves completely differently in presence of free tubulin - it forms tubulin ring stack and promotes de novo microtubule nucleation.
www.biorxiv.org/content/10.1...

We show that human spastin, a microtubule severing enzyme, behaves completely differently in presence of free tubulin - it forms tubulin ring stack and promotes de novo microtubule nucleation.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
#EESMicrotubules will bring together researchers who share an interest in microtubule biology and its relevance to health and disease 🩺
💻 s.embl.org/ees26-09-bl
✒️ Submit your abstract by 11 March

#EESMicrotubules will bring together researchers who share an interest in microtubule biology and its relevance to health and disease 🩺
💻 s.embl.org/ees26-09-bl
✒️ Submit your abstract by 11 March
We report how human outer kinetochore complexes Ndc80 and Ska form cooperative oligomers, that together stabilise microtubule ends against shortening.
www.biorxiv.org/content/10.1...
Key results below: (1/7)
We report how human outer kinetochore complexes Ndc80 and Ska form cooperative oligomers, that together stabilise microtubule ends against shortening.
www.biorxiv.org/content/10.1...
Key results below: (1/7)
SNAP-tag2 for faster and brighter protein labeling
www.nature.com/articles/s41...
Thank you Steffi and Veselin.
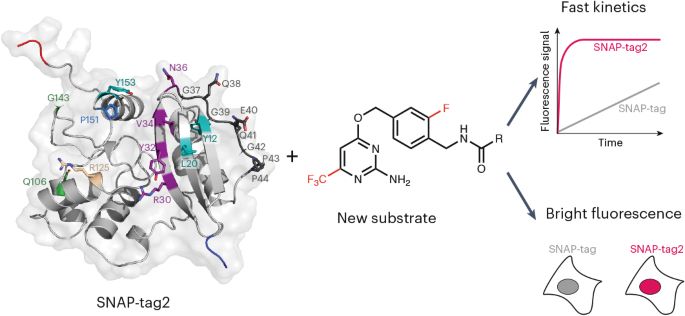
SNAP-tag2 for faster and brighter protein labeling
www.nature.com/articles/s41...
Thank you Steffi and Veselin.
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
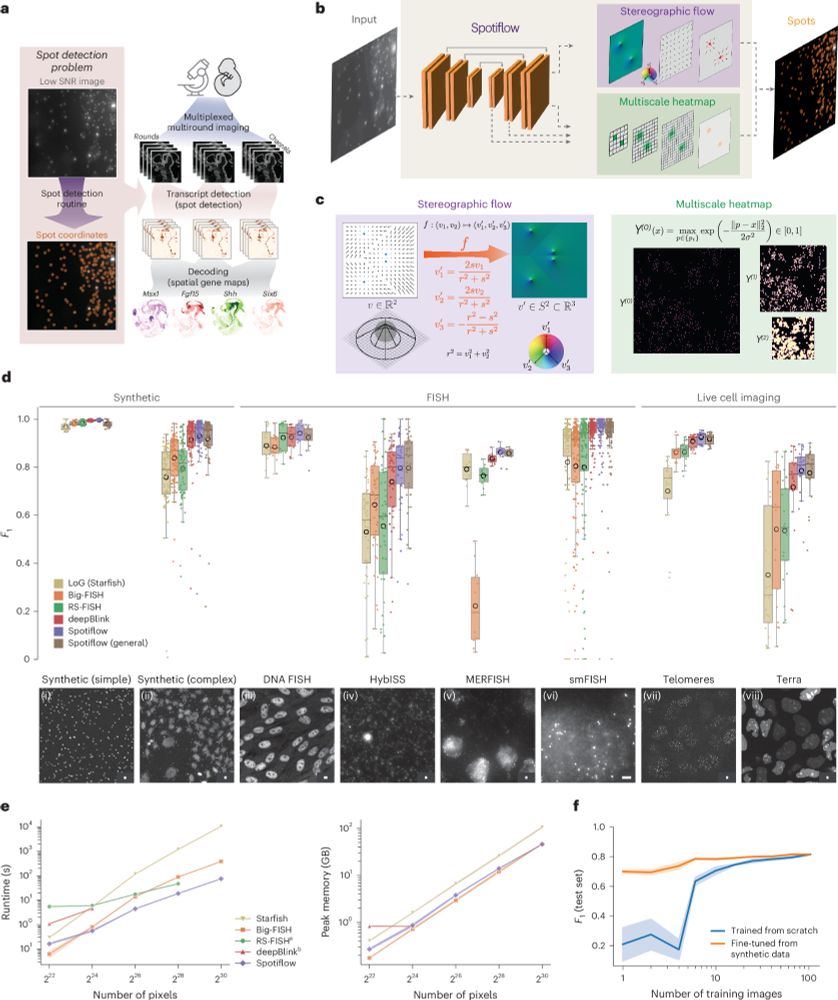
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
First: napari-omero now supports lazy loading of image pyramids from OMERO🔺:
First: napari-omero now supports lazy loading of image pyramids from OMERO🔺:
I just submitted this video to the Nikon Small World in Motion competition. Today is the last day to upload yours! 😉
🧪
I just submitted this video to the Nikon Small World in Motion competition. Today is the last day to upload yours! 😉
🧪


@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
https://napari.org/0.6.0/release/release_0_6_0.html
But to save you a click here's a short thread: 👇
https://napari.org/0.6.0/release/release_0_6_0.html
But to save you a click here's a short thread: 👇
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
We’re thrilled to announce the launch of openRxiv as an independent, researcher-led nonprofit to oversee bioRxiv and medRxiv, the world’s leading preprint servers for life and health sciences.
openrxiv.org/introducing-...
#openRxiv #OpenScience #Preprints #bioRxiv #medRxiv

We’re thrilled to announce the launch of openRxiv as an independent, researcher-led nonprofit to oversee bioRxiv and medRxiv, the world’s leading preprint servers for life and health sciences.
openrxiv.org/introducing-...
#openRxiv #OpenScience #Preprints #bioRxiv #medRxiv
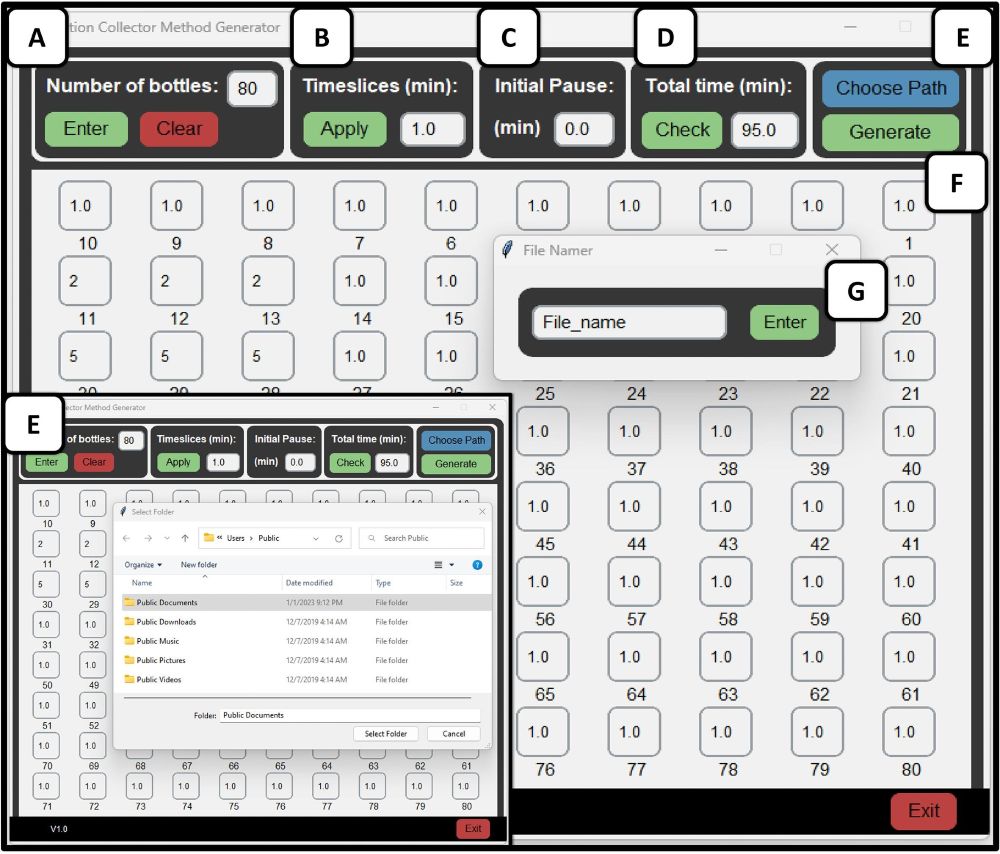
- based on #Creality #Ender3Pro
- cost < US $280
- prep scale fractions: up to 470 mL
- #Python, #GUI, no-code
doi.org/10.1016/j.oh...
#DIYbio #lab #instruments #LC #fluidics #chemistry #DrugDiscovery
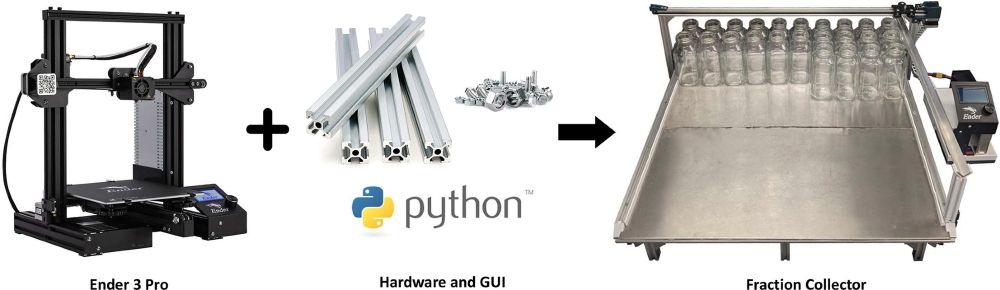

- based on #Creality #Ender3Pro
- cost < US $280
- prep scale fractions: up to 470 mL
- #Python, #GUI, no-code
doi.org/10.1016/j.oh...
#DIYbio #lab #instruments #LC #fluidics #chemistry #DrugDiscovery
forum.image.sc/t/fiji-2024-...
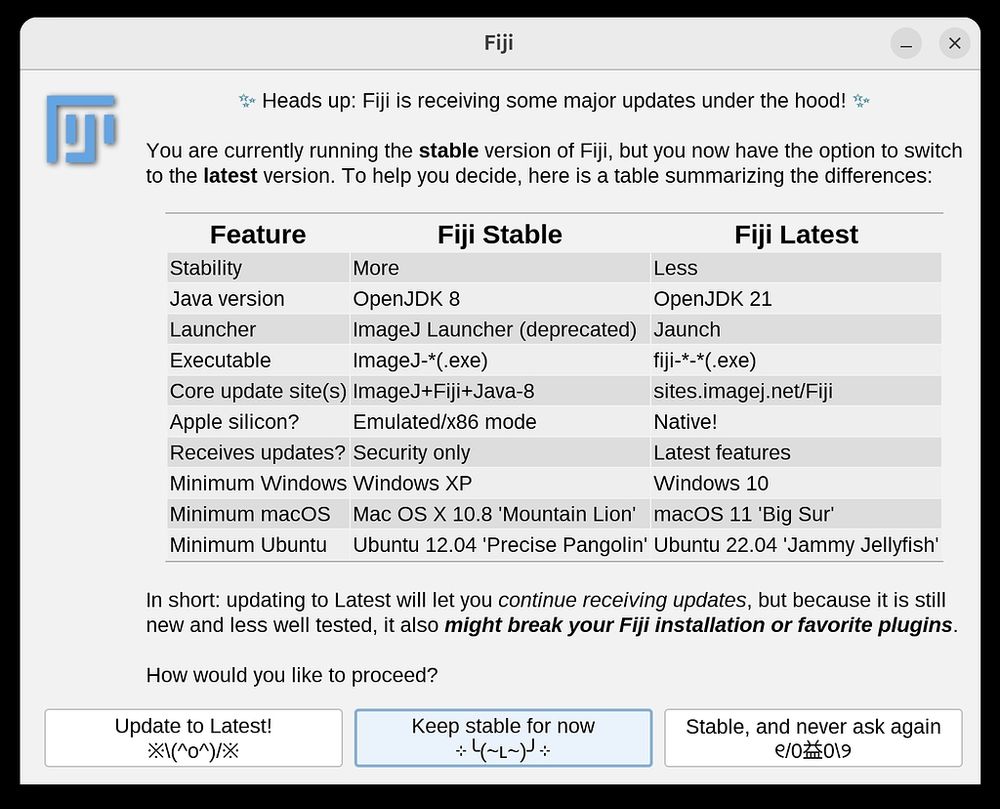
forum.image.sc/t/fiji-2024-...