Manolis Maragkakis
@manolis-maragkakis.bsky.social
2.8K followers
2.3K following
47 posts
Stadtman Principal Investigator @NIH / NIA. Computational Genomics, RNA, Aging, Stress Response, Senescence. Posts are my own.
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Manolis Maragkakis
Olivier Duss
@olivierduss.bsky.social
· Apr 29

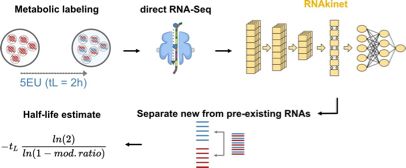
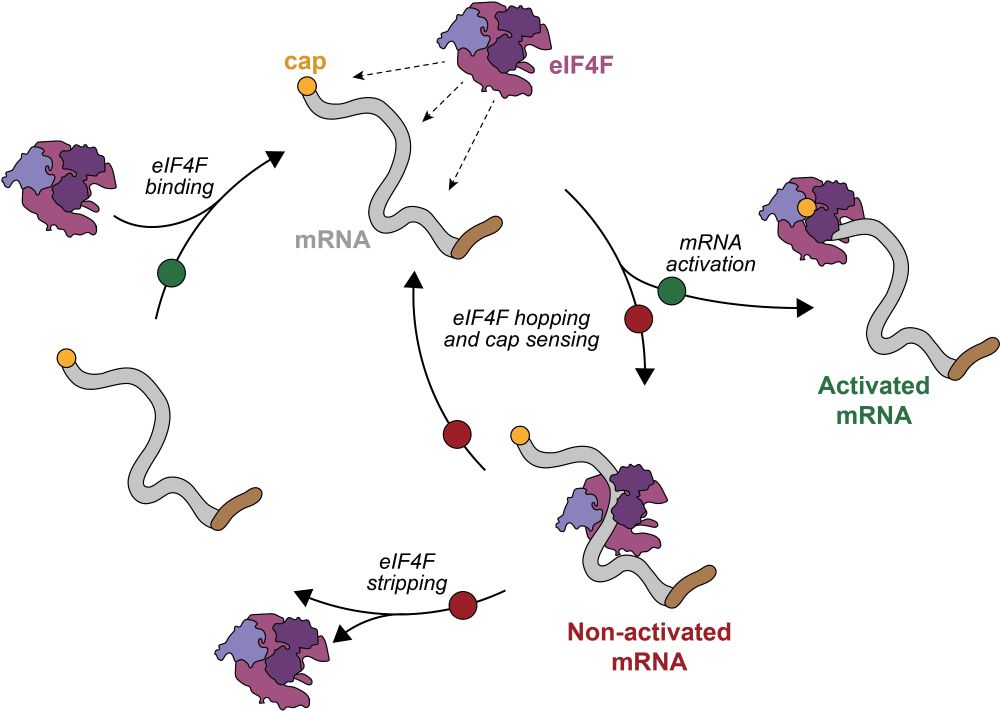
Single-molecule multimodal timing of in vivo mRNA synthesis
mRNA synthesis requires extensive pre-mRNA maturation, the organisation of which remains unclear. Here, we directly sequence pre-mRNA without metabolic labelling or amplification to resolve transcript...
www.biorxiv.org
Reposted by Manolis Maragkakis
Molecular Cell
@cp-molcell.bsky.social
· Apr 10
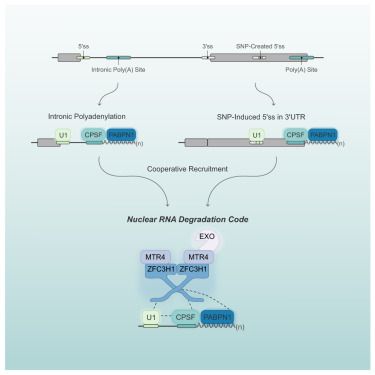
A nuclear RNA degradation code is recognized by PAXT for eukaryotic transcriptome surveillance
Soles et al. demonstrate that the combination of a 5′ splice site and a poly(A) junction constitutes a nuclear RNA degradation code that targets RNAs for degradation by the PAXT adaptor and the RNA exosome. This study provides fundamental insight into how the RNA exosome specifically recognizes misprocessed RNAs.
dlvr.it
Reposted by Manolis Maragkakis
Liam Faller
@liamfaller.bsky.social
· Mar 6
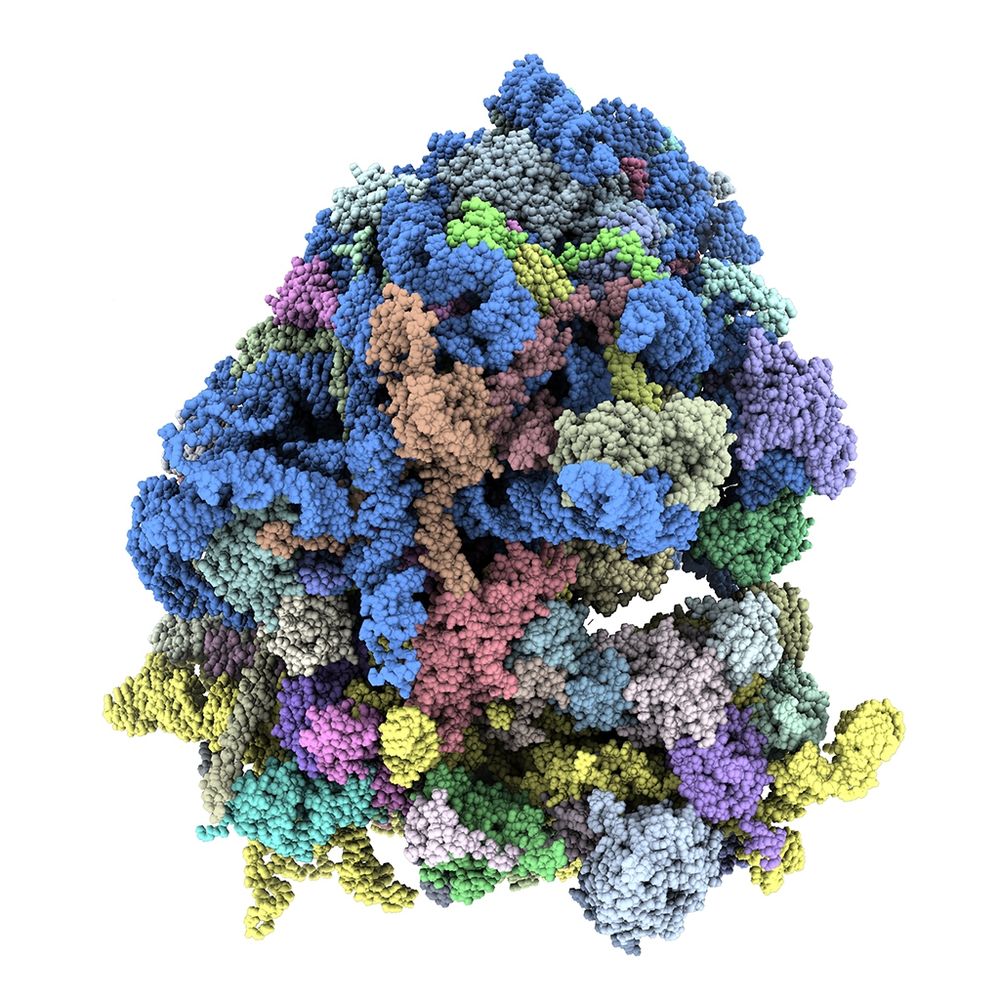
High-throughput approaches for the identification of ribosome heterogeneity | Philosophical Transactions of the Royal Society B: Biological Sciences
Recent advances in the fields of RNA translation and ribosome biology have demonstrated
the heterogeneous nature of ribosomes. This manifests not only across different cellular
contexts but also withi...
royalsocietypublishing.org
Reposted by Manolis Maragkakis
Eugene Valkov
@eugenevalkov.bsky.social
· Feb 12
Molecular Cell
@cp-molcell.bsky.social
· Feb 12

The integrated stress response regulates 18S nonfunctional rRNA decay in mammals
To maintain translational fidelity, cells degrade decoding-incompetent ribosomes through the 18S nonfunctional rRNA decay (NRD) pathway. Coria et al. demonstrate that the concerted actions of GCN2, RNF10, and RIOK3 lead to a block in translation…
dlvr.it
Reposted by Manolis Maragkakis
Reposted by Manolis Maragkakis
Altmetric
@altmetric.com
· Dec 11
Reposted by Manolis Maragkakis
Reposted by Manolis Maragkakis
Reposted by Manolis Maragkakis