Matt Girgenti
@mattgirgenti.bsky.social
80 followers
81 following
13 posts
Assistant Prof YalePsych and DeptVetAffairs. Single-cell, Genomics, Neuroscience, Psychiatric Genetics. Dad of 2. MtG collector. Opinions are mine
Posts
Media
Videos
Starter Packs
Pinned
Matt Girgenti
@mattgirgenti.bsky.social
· Jun 18
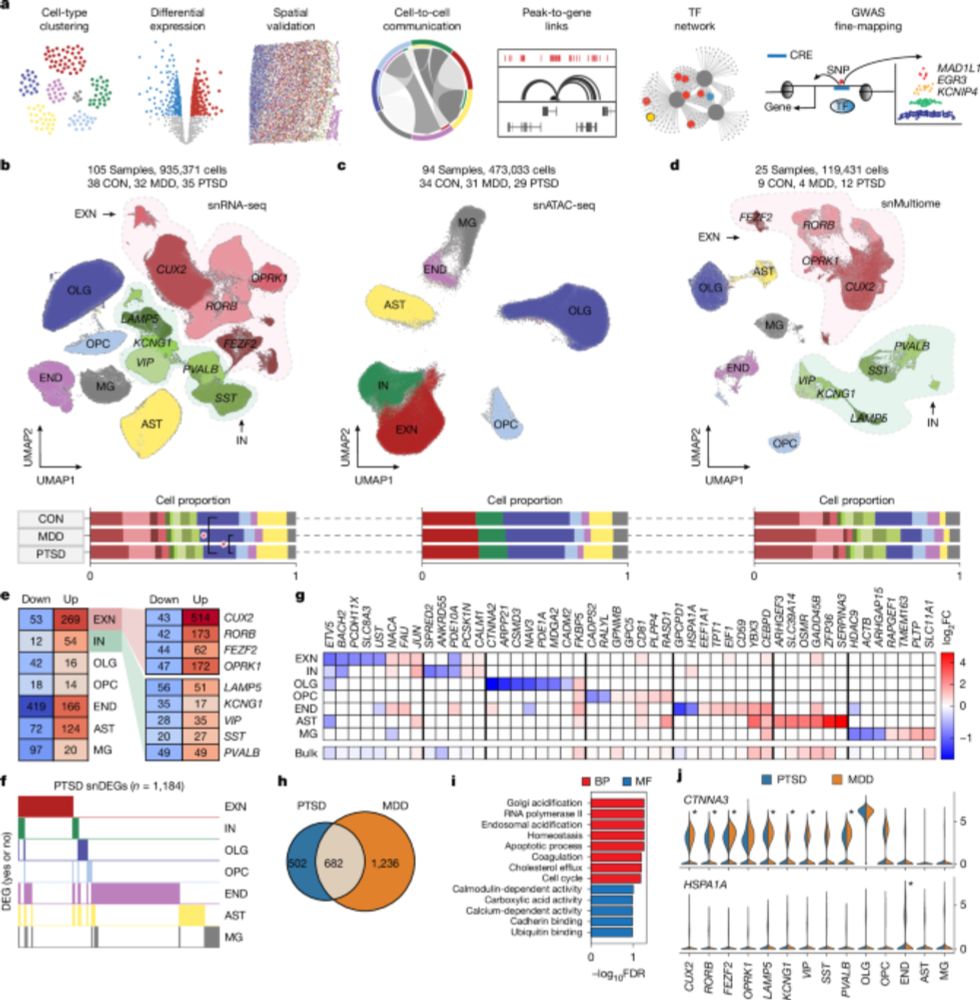
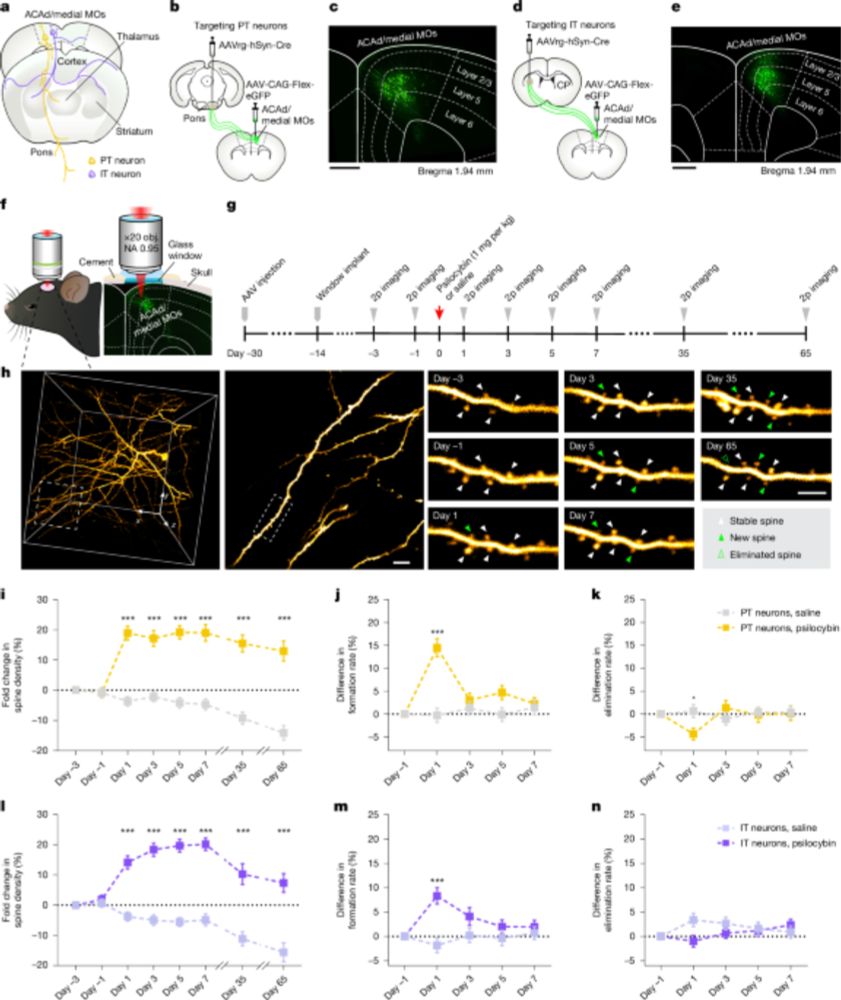
Single-cell transcriptomic and chromatin dynamics of the human brain in PTSD - Nature
A comprehensive analysis of the cell-specific molecular regulatory mechanisms underlying post-traumatic stress disorder in the human prefrontal cortex.
www.nature.com
Reposted by Matt Girgenti
Reposted by Matt Girgenti
Jeremy Day
@daylab.bsky.social
· Aug 15
Reposted by Matt Girgenti
Reposted by Matt Girgenti
Matt Girgenti
@mattgirgenti.bsky.social
· Jun 18
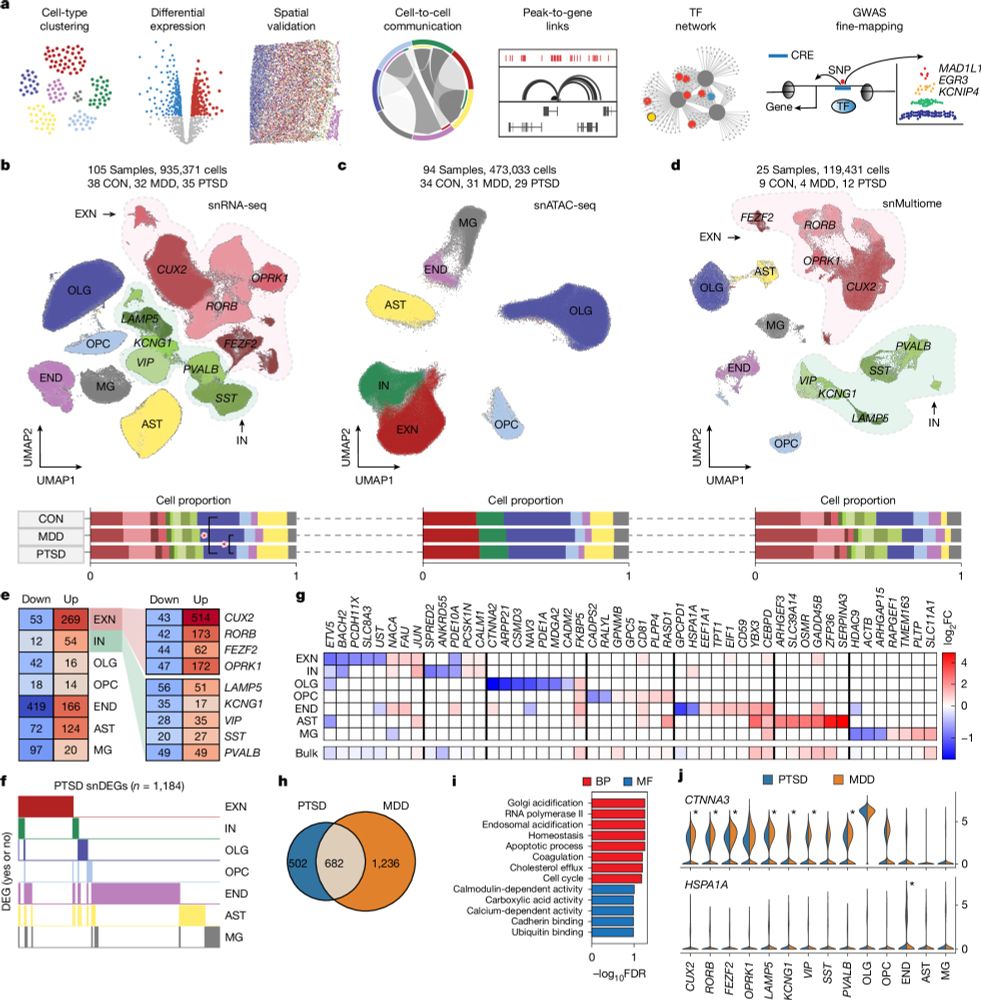
Single-cell transcriptomic and chromatin dynamics of the human brain in PTSD
Nature - A comprehensive analysis of the cell-specific molecular regulatory mechanisms underlying post-traumatic stress disorder in the human prefrontal cortex.
nam12.safelinks.protection.outlook.com
Matt Girgenti
@mattgirgenti.bsky.social
· Jun 18
Matt Girgenti
@mattgirgenti.bsky.social
· Jun 18
Matt Girgenti
@mattgirgenti.bsky.social
· Jun 18

Single-cell transcriptomic and chromatin dynamics of the human brain in PTSD - Nature
A comprehensive analysis of the cell-specific molecular regulatory mechanisms underlying post-traumatic stress disorder in the human prefrontal cortex.
www.nature.com
Reposted by Matt Girgenti
Reposted by Matt Girgenti
Matt Girgenti
@mattgirgenti.bsky.social
· Apr 30
Reposted by Matt Girgenti
Reposted by Matt Girgenti
Reposted by Matt Girgenti