Matt Howard
@matthewkhoward.bsky.social
150 followers
250 following
23 posts
Doing science @UCSF in the Coyote-Maestas and Manglik Labs. Former Jackrel Lab @WUSTL www.matthewkhoward.com
Posts
Media
Videos
Starter Packs
Pinned
Matt Howard
@matthewkhoward.bsky.social
· Jul 21
Matt Howard
@matthewkhoward.bsky.social
· Jun 30
Jerome
@jeromics.bsky.social
· Jun 30

Accurate variant effect estimation in FACS-based deep mutational scanning data with Lilace
Deep mutational scanning (DMS) experiments interrogate the effect of genetic variants on protein function, often using fluorescence-activated cell sorting (FACS) to quantitatively measure molecular ph...
www.biorxiv.org
Reposted by Matt Howard
Ishan Deshpande
@ishand.bsky.social
· Mar 26

Postdoctoral Fellow - Structural Biology, Deshpande Lab in South San Francisco, California, United States of America | Students & Graduates at Genentech
Apply for Postdoctoral Fellow - Structural Biology, Deshpande Lab job with Genentech in South San Francisco, California, United States of America. Students & Graduates at Genentech
careers.gene.com
Reposted by Matt Howard
Vijay Ramani
@vram142.bsky.social
· Mar 26

Structures of the PI3Kα/KRas complex on lipid bilayers reveal the molecular mechanism of PI3Kα activation
PI3Kα is a potent oncogene that converts PIP2 to PIP3 at the plasma membrane upon activation by receptor tyrosine kinases and Ras GTPases. In the absence of any structures of activated PI3Kα, the mole...
www.biorxiv.org
Reposted by Matt Howard
Reposted by Matt Howard
Emily Blythe
@emilyblythe.bsky.social
· Feb 27

Endocytosis sculpts distinct cAMP signal transduction by endogenously coexpressed GPCRs
Many G protein-coupled receptors (GPCRs) trigger a second phase of G protein-dependent signaling from internal membranes after agonist-induced endocytosis. However, individual GPCRs differ significant...
www.biorxiv.org
Reposted by Matt Howard
Vijay Ramani
@vram142.bsky.social
· Jan 2
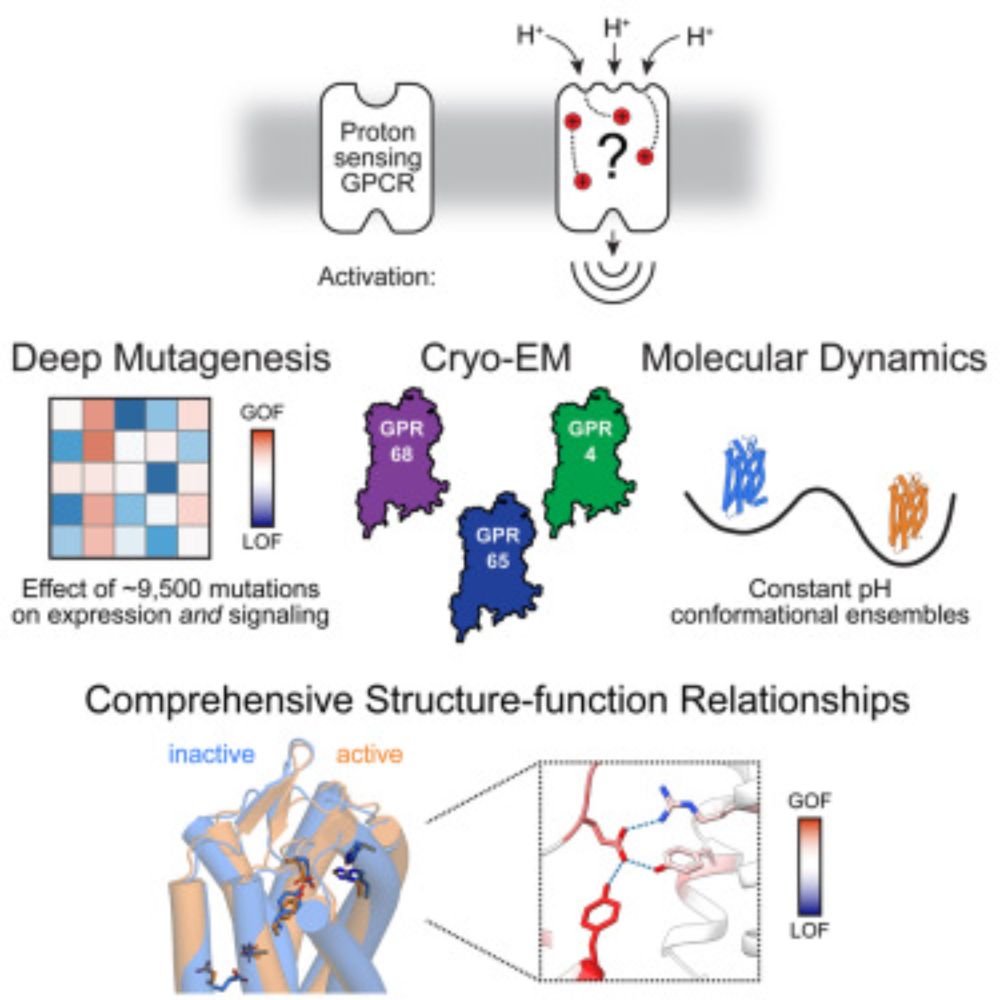
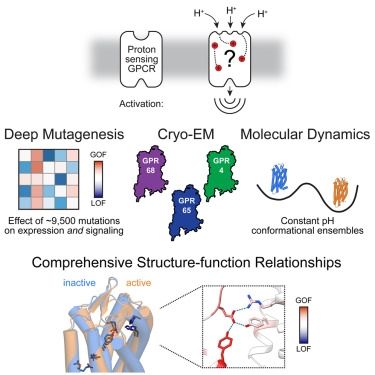
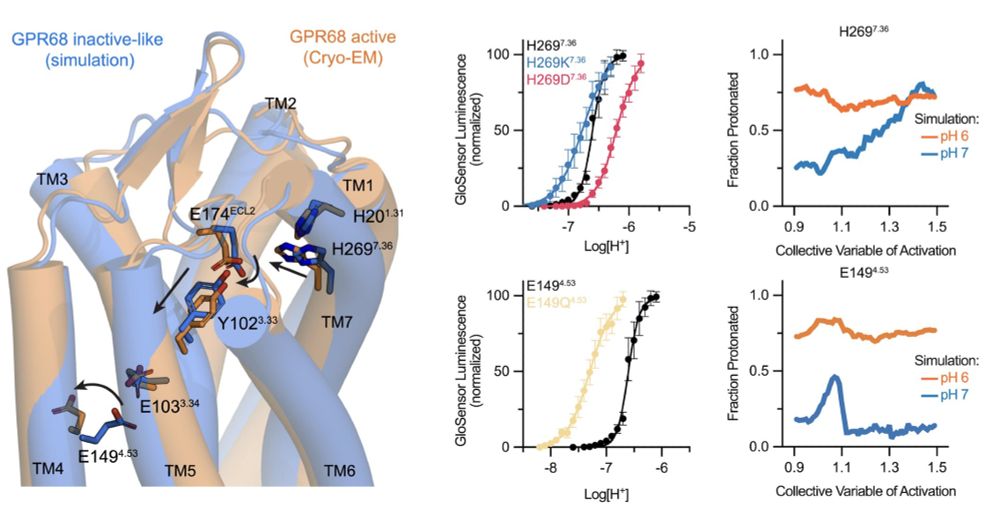
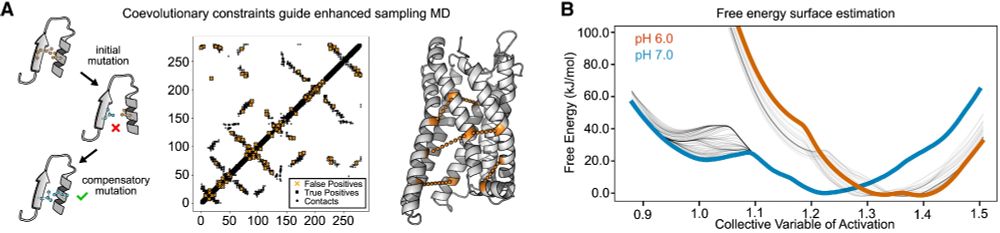
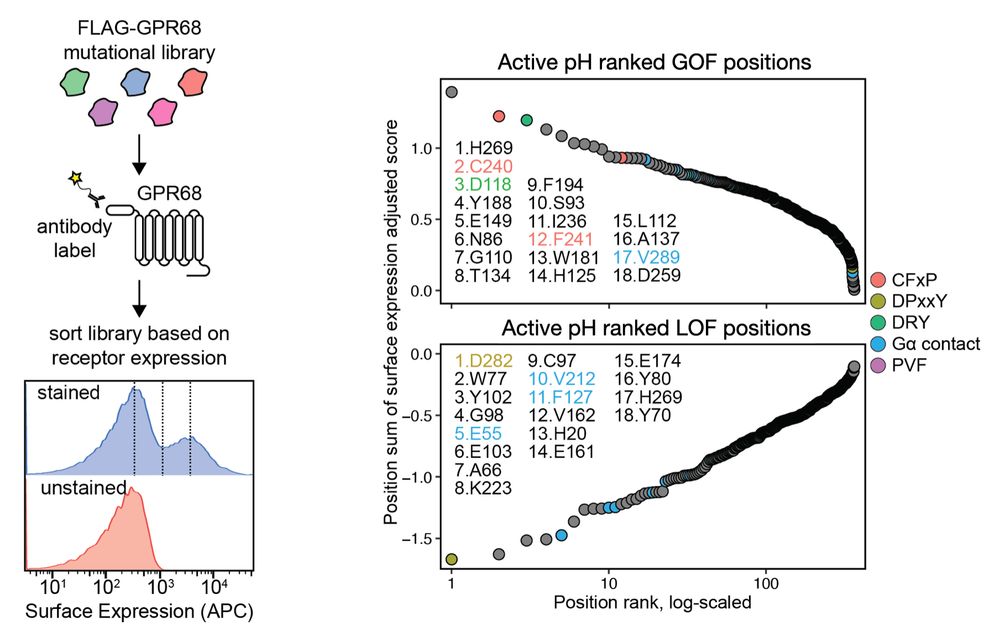
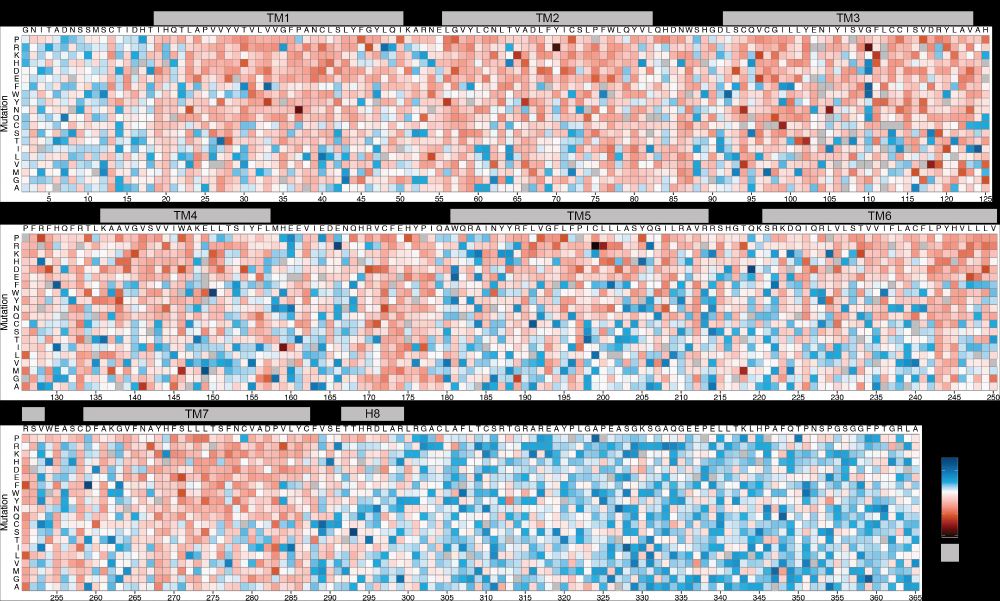
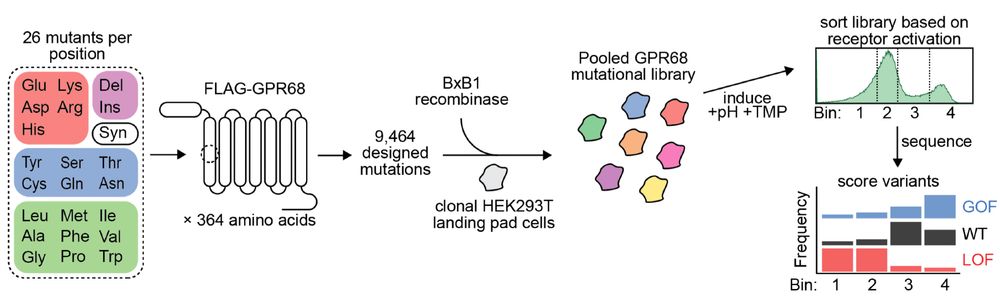
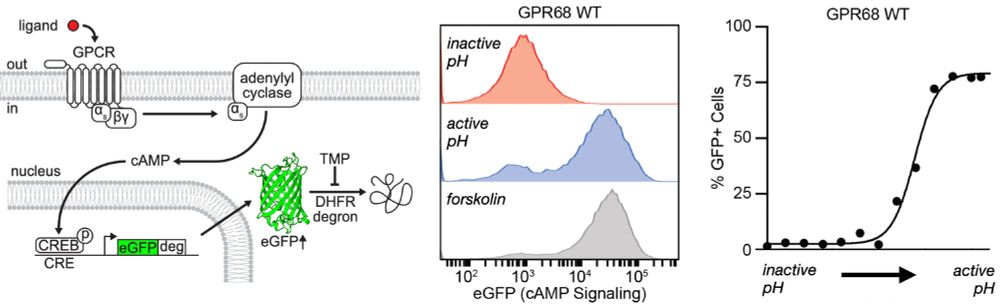
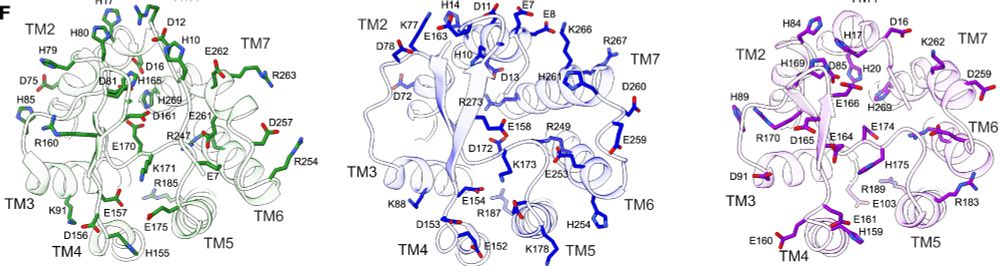
Molecular basis of proton sensing by G protein-coupled receptors
Howard et al. combine mechanistic deep mutational scanning, cryo-EM, and constant-pH molecular dynamics simulations to provide a holistic view of proton activation in human pH-sensitive GPCRs.
www.cell.com