
Matthias Meyer-Bender
@matthiasmeybe.bsky.social
PhD student at EMBL Heidelberg
Interested in computational biology, biological image analysis and AI in medicine
Interested in computational biology, biological image analysis and AI in medicine
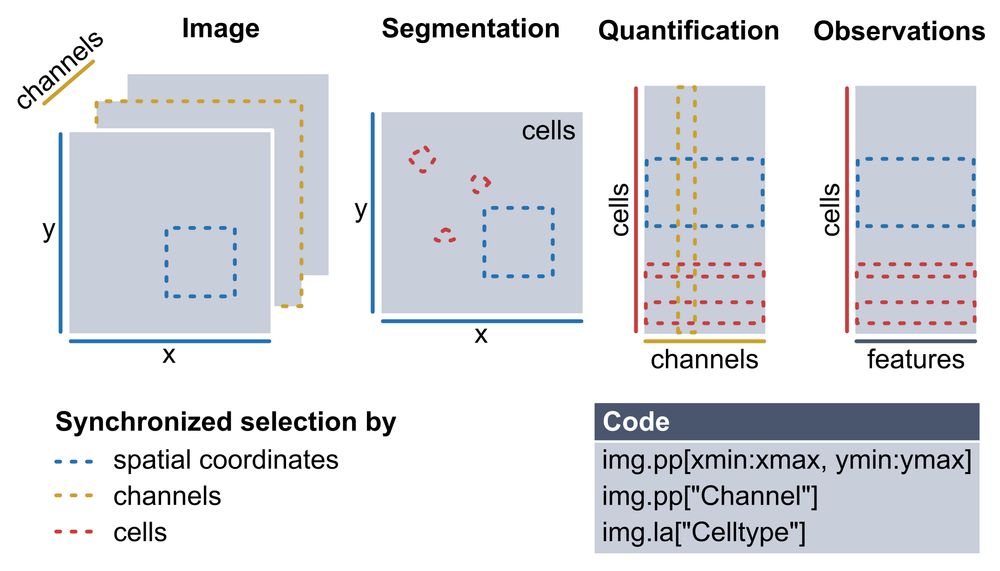
Multiplexed imaging (CODEX, MICS, IMC) gives single-cell resolution at the protein level — but analyzing these datasets requires stitching together many different tools and data structures.
You need to manage images, masks, expression matrices, and keep them all consistent.
You need to manage images, masks, expression matrices, and keep them all consistent.
May 5, 2025 at 11:30 AM
Multiplexed imaging (CODEX, MICS, IMC) gives single-cell resolution at the protein level — but analyzing these datasets requires stitching together many different tools and data structures.
You need to manage images, masks, expression matrices, and keep them all consistent.
You need to manage images, masks, expression matrices, and keep them all consistent.
New preprint out!
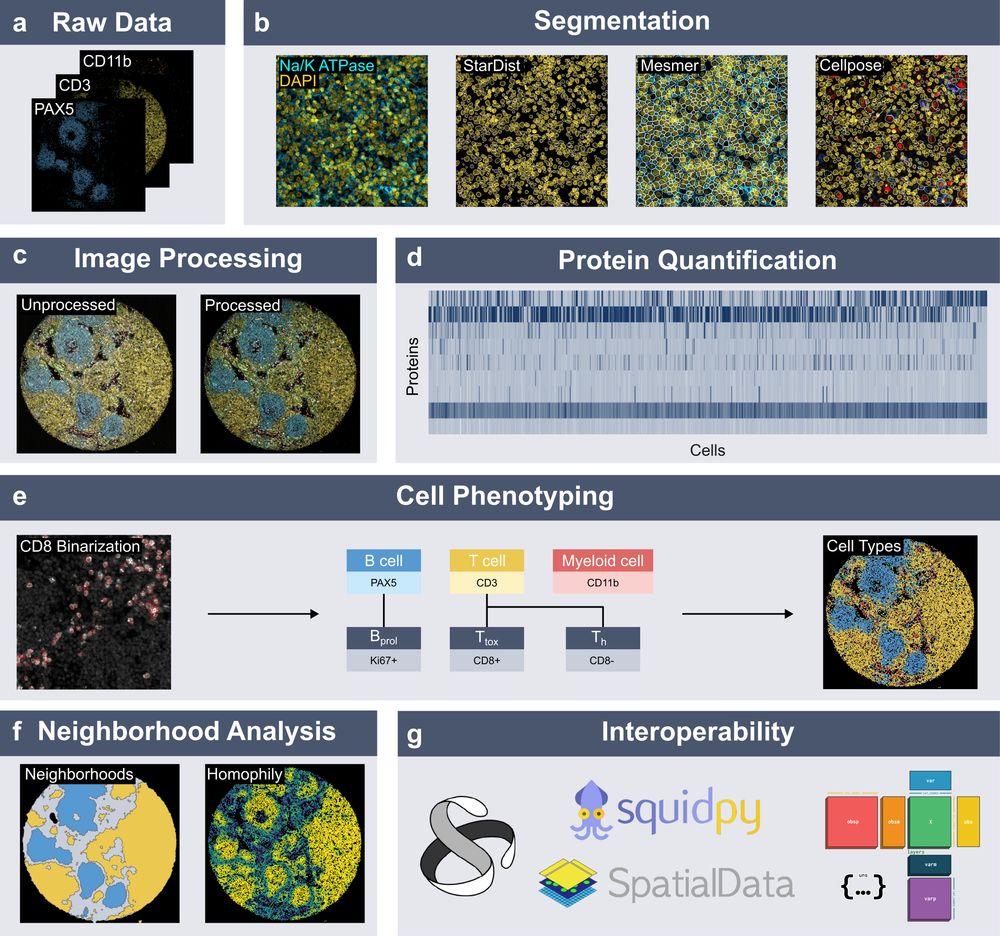
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
May 5, 2025 at 11:30 AM
New preprint out!
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...

