
Matthias Meyer-Bender
@matthiasmeybe.bsky.social
PhD student at EMBL Heidelberg
Interested in computational biology, biological image analysis and AI in medicine
Interested in computational biology, biological image analysis and AI in medicine
Pinned
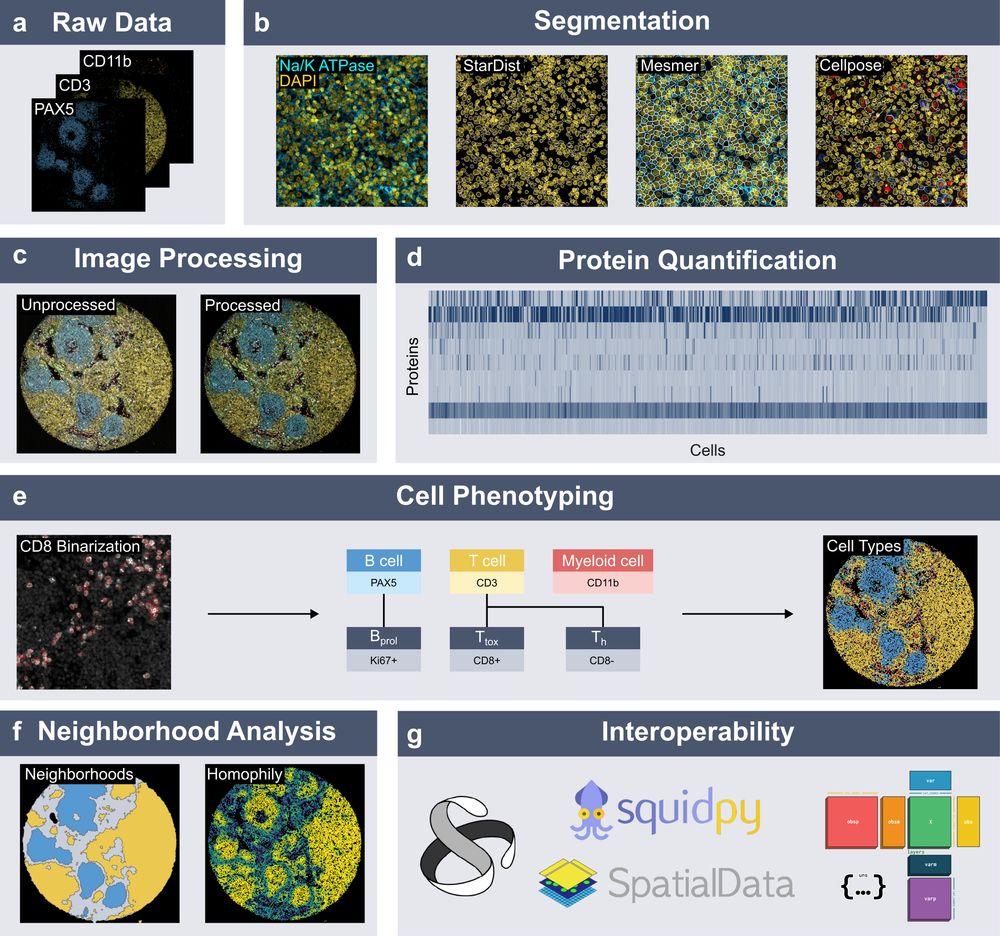
New preprint out!
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
Reposted by Matthias Meyer-Bender
TLDR; The PSF has made the decision to put our community and our shared diversity, equity, and inclusion values ahead of seeking $1.5M in new revenue. Please read and share. pyfound.blogspot.com/2025/10/NSF-...
🧵
🧵

The official home of the Python Programming Language
www.python.org
October 27, 2025 at 2:47 PM
TLDR; The PSF has made the decision to put our community and our shared diversity, equity, and inclusion values ahead of seeking $1.5M in new revenue. Please read and share. pyfound.blogspot.com/2025/10/NSF-...
🧵
🧵
Reposted by Matthias Meyer-Bender
Latitudinal diversity gradients (LDGs) are found across 🌱🐨🦠 but their underlying mechanisms remain unclear.
In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...
In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...

Variations in the latitudinal diversity gradients of the ocean microbiome
Latitudinal diversity gradients (LDGs), typically declining from the equator to the poles, are among the most pervasive macroecological patterns, yet their generality and underlying drivers in the oce...
doi.org
October 15, 2025 at 12:05 PM
Latitudinal diversity gradients (LDGs) are found across 🌱🐨🦠 but their underlying mechanisms remain unclear.
In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...
In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...
Reposted by Matthias Meyer-Bender
(Huber lab)[EMBL] welcomes applications for a PhD in ML for spatial omics [representation learning + integration with biostatistics, cell + anatomy foundation models, collaborate with domain scientists on cancer and dev:bio discovery science] |> Apply though the ELLIS portal ::: Deadline 2025-10-31
🎓 The application portal for the #ELLISPhD Program is open today! Info & link: https://bit.ly/45DSe75

ELLIS PhD Program: Call for Applications 2025
The ELLIS mission is to create a diverse European network that promotes research excellence and advances breakthroughs in AI, as well as a pan-European PhD program to educate the next generation of AI...
ellis.eu
October 4, 2025 at 8:31 AM
(Huber lab)[EMBL] welcomes applications for a PhD in ML for spatial omics [representation learning + integration with biostatistics, cell + anatomy foundation models, collaborate with domain scientists on cancer and dev:bio discovery science] |> Apply though the ELLIS portal ::: Deadline 2025-10-31
Reposted by Matthias Meyer-Bender
Happy to share that ShapeEmbed has been accepted at @neuripsconf.bsky.social 🎉 SE is self-supervised framework to encode 2D contours from microscopy & natural images into a latent representation invariant to translation, scaling, rotation, reflection & point indexing
📄 arxiv.org/pdf/2507.01009 (1/N)
📄 arxiv.org/pdf/2507.01009 (1/N)
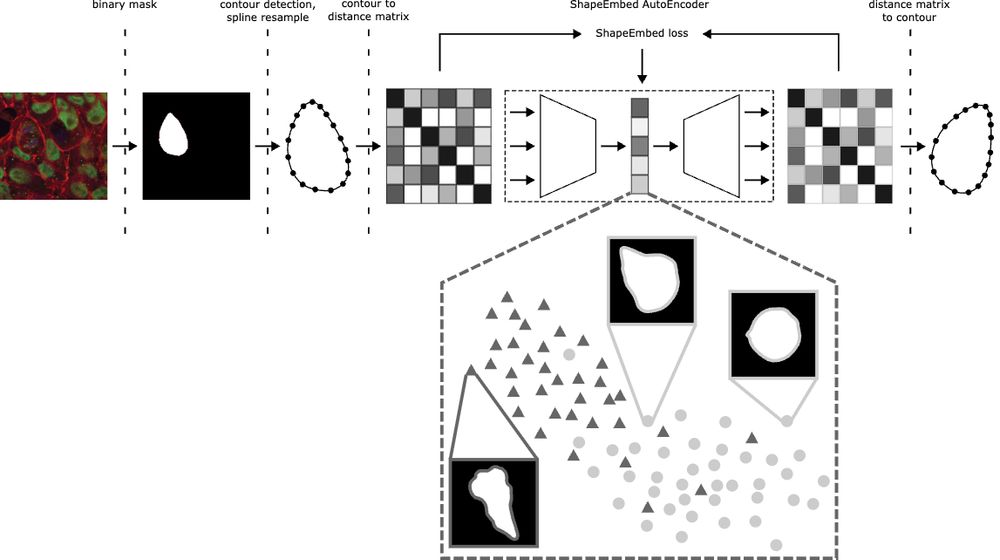
September 23, 2025 at 8:32 AM
Happy to share that ShapeEmbed has been accepted at @neuripsconf.bsky.social 🎉 SE is self-supervised framework to encode 2D contours from microscopy & natural images into a latent representation invariant to translation, scaling, rotation, reflection & point indexing
📄 arxiv.org/pdf/2507.01009 (1/N)
📄 arxiv.org/pdf/2507.01009 (1/N)
Reposted by Matthias Meyer-Bender
EMBL International PhD Programme - winter recruitment 2026! 📣
Research groups across EMBL are recruiting now! www.embl.org/about/info/e...
Don’t miss this opportunity to receive dedicated mentoring while doing interdisciplinary research.
Research groups across EMBL are recruiting now! www.embl.org/about/info/e...
Don’t miss this opportunity to receive dedicated mentoring while doing interdisciplinary research.

September 22, 2025 at 11:42 AM
EMBL International PhD Programme - winter recruitment 2026! 📣
Research groups across EMBL are recruiting now! www.embl.org/about/info/e...
Don’t miss this opportunity to receive dedicated mentoring while doing interdisciplinary research.
Research groups across EMBL are recruiting now! www.embl.org/about/info/e...
Don’t miss this opportunity to receive dedicated mentoring while doing interdisciplinary research.
Reposted by Matthias Meyer-Bender
We are excited to share the publication of our paper on exploratory spatial statistics for spatial omics data
academic.oup.com/nar/article/...
academic.oup.com/nar/article/...

Harnessing the potential of spatial statistics for spatial omics data with pasta
Abstract. Spatial omics allow for the molecular characterization of cells in their spatial context. Notably, the two main technological streams, imaging-ba
academic.oup.com
September 11, 2025 at 7:04 AM
We are excited to share the publication of our paper on exploratory spatial statistics for spatial omics data
academic.oup.com/nar/article/...
academic.oup.com/nar/article/...
Reposted by Matthias Meyer-Bender
Our paper benchmarking foundation models for perturbation effect prediction is finally published 🎉🥳🎉
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵


August 4, 2025 at 1:52 PM
Our paper benchmarking foundation models for perturbation effect prediction is finally published 🎉🥳🎉
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
Reposted by Matthias Meyer-Bender
Update: We greatly revised our paper and renamed it “Harnessing the Potential of Spatial Statistics for Spatial Omics Data with pasta”.
We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561
We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561

Harnessing the Potential of Spatial Statistics for Spatial Omics Data with pasta
Spatial omics assays allow for the molecular characterisation of cells in their spatial context. Notably, the two main technological streams, imaging-based and high-throughput sequencing-based, can gi...
arxiv.org
June 27, 2025 at 1:28 PM
Update: We greatly revised our paper and renamed it “Harnessing the Potential of Spatial Statistics for Spatial Omics Data with pasta”.
We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561
We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561
Reposted by Matthias Meyer-Bender
Very happy and proud to announce that the first preprint of my PhD is out: arxiv.org/abs/2504.20710
We developed an R package to translate mathematical models in SBML format into executable odin models and visualise models from @biomodels.bsky.social on our website Menelmacar biomodels.bacpop.org
We developed an R package to translate mathematical models in SBML format into executable odin models and visualise models from @biomodels.bsky.social on our website Menelmacar biomodels.bacpop.org

SBMLtoOdin and Menelmacar: Interactive visualisation of systems biology models for expert and non-expert audiences
Motivation: Computational models in biology can increase our understanding of biological systems, be used to answer research questions, and make predictions. Accessibility and reusability of computati...
arxiv.org
May 7, 2025 at 9:27 AM
Very happy and proud to announce that the first preprint of my PhD is out: arxiv.org/abs/2504.20710
We developed an R package to translate mathematical models in SBML format into executable odin models and visualise models from @biomodels.bsky.social on our website Menelmacar biomodels.bacpop.org
We developed an R package to translate mathematical models in SBML format into executable odin models and visualise models from @biomodels.bsky.social on our website Menelmacar biomodels.bacpop.org
New preprint out!
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...

May 5, 2025 at 11:30 AM
New preprint out!
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
Reposted by Matthias Meyer-Bender
Spatialproteomics - an interoperable toolbox for analyzing highly multiplexed fluorescence image data https://www.biorxiv.org/content/10.1101/2025.04.29.651202v1
May 3, 2025 at 11:47 PM
Spatialproteomics - an interoperable toolbox for analyzing highly multiplexed fluorescence image data https://www.biorxiv.org/content/10.1101/2025.04.29.651202v1
Reposted by Matthias Meyer-Bender
Postdoc - Statistics and machine learning with spatial omics and single-cell perturbation data
www.huber.embl.de/group/posts/...
www.huber.embl.de/group/posts/...
Postdoc - Statistics and machine learning with spatial omics and single-cell perturbation data – Huber Group @ EMBL
Precision Oncology and Multiomics
www.huber.embl.de
March 12, 2025 at 1:08 PM
Postdoc - Statistics and machine learning with spatial omics and single-cell perturbation data
www.huber.embl.de/group/posts/...
www.huber.embl.de/group/posts/...
Reposted by Matthias Meyer-Bender
*Medical Data Scientist Postdoc* Program by Medical Faculty Uni Heidelberg. Join S.Dietrich, J.Lu & me to work on statistical& AI methods applied to single cell and spatial omics to improve immunotherapies:
www.medizinische-fakultaet-hd.uni-heidelberg.de/forschung/fo...
www.embl.org/about/info/m...
www.medizinische-fakultaet-hd.uni-heidelberg.de/forschung/fo...
www.embl.org/about/info/m...
Medizinische Fakultät Heidelberg: Medical Data Scientist Programm
www.medizinische-fakultaet-hd.uni-heidelberg.de
October 3, 2023 at 12:59 PM
*Medical Data Scientist Postdoc* Program by Medical Faculty Uni Heidelberg. Join S.Dietrich, J.Lu & me to work on statistical& AI methods applied to single cell and spatial omics to improve immunotherapies:
www.medizinische-fakultaet-hd.uni-heidelberg.de/forschung/fo...
www.embl.org/about/info/m...
www.medizinische-fakultaet-hd.uni-heidelberg.de/forschung/fo...
www.embl.org/about/info/m...

