
Matthias Meyer-Bender
@matthiasmeybe.bsky.social
PhD student at EMBL Heidelberg
Interested in computational biology, biological image analysis and AI in medicine
Interested in computational biology, biological image analysis and AI in medicine
#spatialproteomics #spatialbiology #multiplexedimaging #bioinformatics #python #scverse #opensource #singlecell #akoya #codex
May 5, 2025 at 11:30 AM
It integrates with spatialdata and anndata, helping extend scverse toward spatial proteomics and imaging workflows.
We hope it helps researchers build flexible, powerful analysis pipelines for analyzing highly multiplexed fluorescence images.
🔧 Package: github.com/sagar87/spat...
We hope it helps researchers build flexible, powerful analysis pipelines for analyzing highly multiplexed fluorescence images.
🔧 Package: github.com/sagar87/spat...

GitHub - sagar87/spatialproteomics: Spatialproteomics is a light weight wrapper around xarray with the intention to facilitate the data exploration and analysis of highly multiplexed immunohistochemistry data. Docs available here: https://sagar87.github.io/spatialproteomics/ .
Spatialproteomics is a light weight wrapper around xarray with the intention to facilitate the data exploration and analysis of highly multiplexed immunohistochemistry data. Docs available here: h...
github.com
May 5, 2025 at 11:30 AM
It integrates with spatialdata and anndata, helping extend scverse toward spatial proteomics and imaging workflows.
We hope it helps researchers build flexible, powerful analysis pipelines for analyzing highly multiplexed fluorescence images.
🔧 Package: github.com/sagar87/spat...
We hope it helps researchers build flexible, powerful analysis pipelines for analyzing highly multiplexed fluorescence images.
🔧 Package: github.com/sagar87/spat...
𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬 offers:
✅ A unified API for segmentation, image processing, cell phenotyping, and spatial statistics
✅Consistent handling of shared dimensions across data structures
✅Built on xarray and dask for high flexibility and memory efficiency
✅Easy installation and usage
✅ A unified API for segmentation, image processing, cell phenotyping, and spatial statistics
✅Consistent handling of shared dimensions across data structures
✅Built on xarray and dask for high flexibility and memory efficiency
✅Easy installation and usage
May 5, 2025 at 11:30 AM
𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬 offers:
✅ A unified API for segmentation, image processing, cell phenotyping, and spatial statistics
✅Consistent handling of shared dimensions across data structures
✅Built on xarray and dask for high flexibility and memory efficiency
✅Easy installation and usage
✅ A unified API for segmentation, image processing, cell phenotyping, and spatial statistics
✅Consistent handling of shared dimensions across data structures
✅Built on xarray and dask for high flexibility and memory efficiency
✅Easy installation and usage
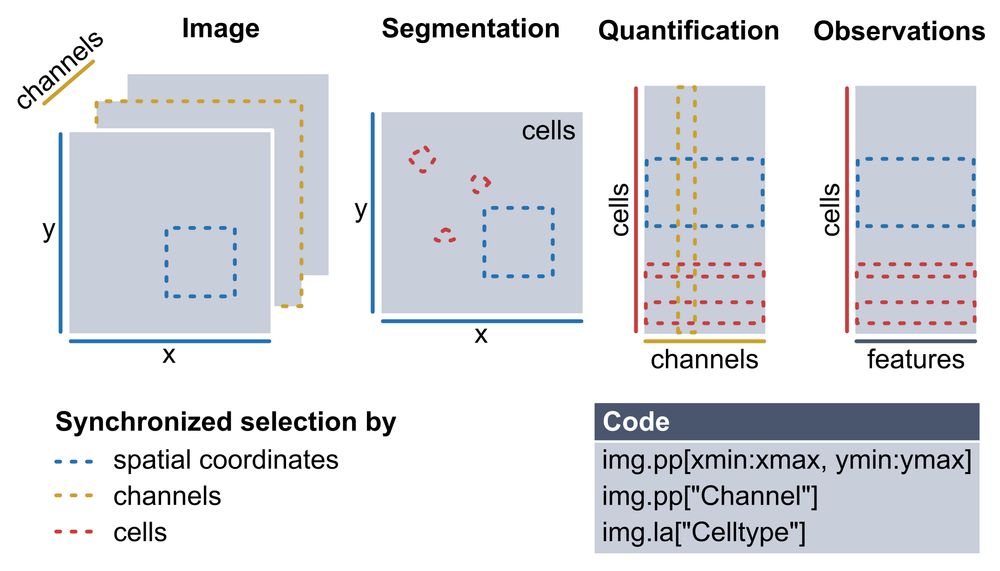
Multiplexed imaging (CODEX, MICS, IMC) gives single-cell resolution at the protein level — but analyzing these datasets requires stitching together many different tools and data structures.
You need to manage images, masks, expression matrices, and keep them all consistent.
You need to manage images, masks, expression matrices, and keep them all consistent.
May 5, 2025 at 11:30 AM
Multiplexed imaging (CODEX, MICS, IMC) gives single-cell resolution at the protein level — but analyzing these datasets requires stitching together many different tools and data structures.
You need to manage images, masks, expression matrices, and keep them all consistent.
You need to manage images, masks, expression matrices, and keep them all consistent.
Mattermost is nice for communication, although it limits the file size, so maybe not ideal for sharing larger files. If you regularly share big files, maybe setting up ownCloud could be an option?
October 10, 2023 at 8:31 PM
Mattermost is nice for communication, although it limits the file size, so maybe not ideal for sharing larger files. If you regularly share big files, maybe setting up ownCloud could be an option?

