Maxim Igaev
@maxotubule.bsky.social
370 followers
1.2K following
10 posts
Computational Biophysicist and Royal Society University Research Fellow at the School of Life Sciences, University of Dundee 🇬🇧
Studying kinetochore-microtubule magic 🪄
Views are my own.
https://www.dundee.ac.uk/people/maxim-igaev
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Maxim Igaev
Reposted by Maxim Igaev
CompBioPhys
@compbiophys.bsky.social
· Jul 2
Reposted by Maxim Igaev
CompBioPhys
@compbiophys.bsky.social
· Jul 2
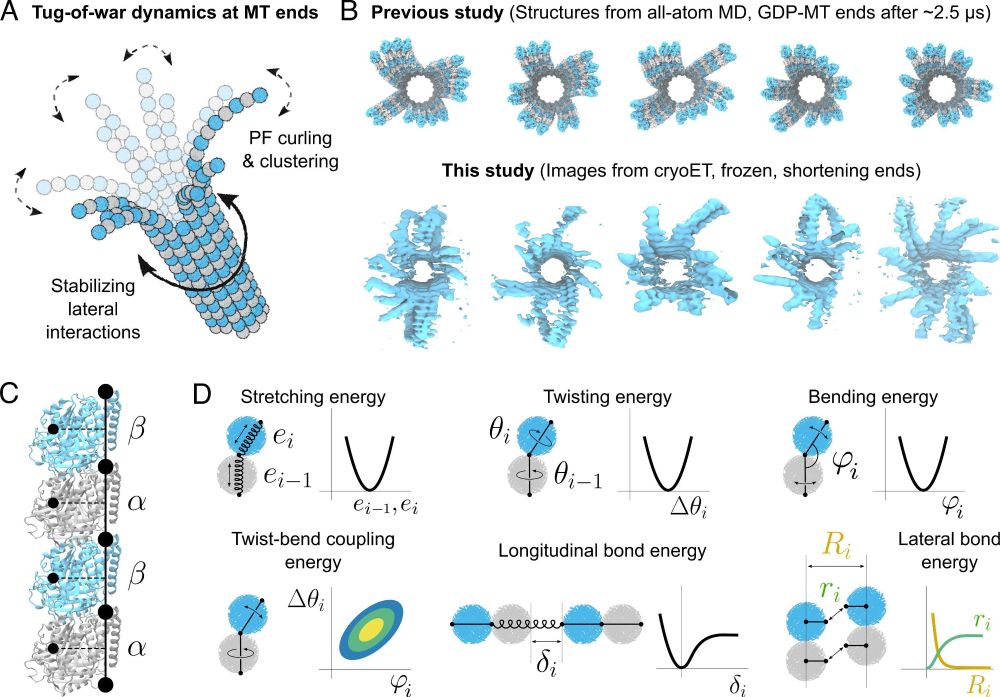
Microtubule dynamics are defined by conformations and stability of clustered protofilaments | PNAS
Microtubules are dynamic cytoskeletal polymers that add and lose tubulin dimers at
their ends. Microtubule growth, shortening, and transitions betw...
www.pnas.org
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Manuel Thery
@manuelthery.bsky.social
· Jun 4
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Maxim Igaev
@maxotubule.bsky.social
· Apr 14
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Maxim Igaev
@maxotubule.bsky.social
· Jan 7
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Michal Kolář
@mhko.bsky.social
· Dec 14
Reposted by Maxim Igaev
Martin Vögele
@martinvoegele.bsky.social
· Dec 13
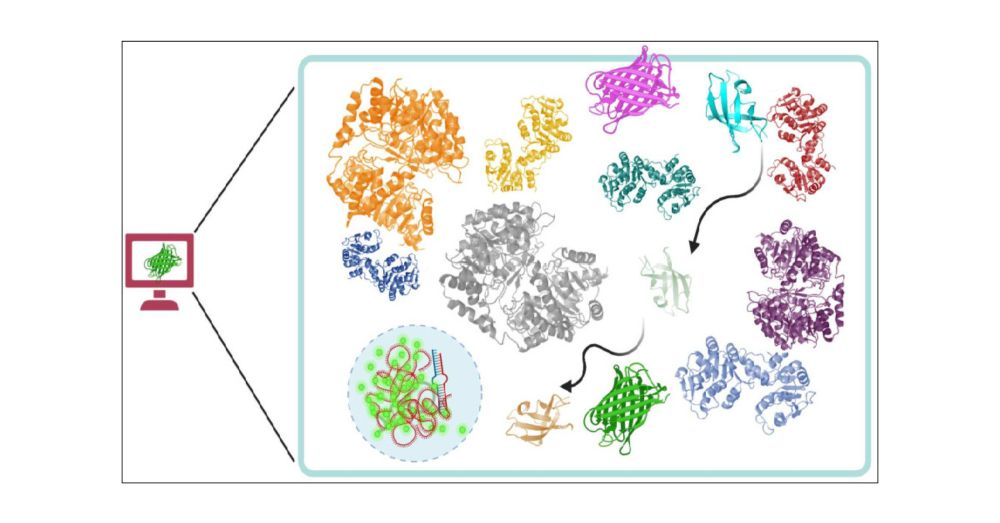
Recent Progress in Modeling and Simulation of Biomolecular Crowding and Condensation Inside Cells
Macromolecular crowding in the cellular cytoplasm can potentially impact diffusion rates of proteins, their intrinsic structural stability, binding of proteins to their corresponding partners as well ...
pubs.acs.org
Reposted by Maxim Igaev
Reposted by Maxim Igaev
Jorge Ferreira
@jorgeferreira.bsky.social
· Nov 13
Reposted by Maxim Igaev
Kirsty Wan
@micromotility.bsky.social
· Nov 13