Meghana Oak
@meghana-oak.bsky.social
58 followers
110 following
8 posts
PhD at IES @www.helmholtz-munich.de | BS-MS @IISER Kolkata
Epigenetics, embryonic development, cellular reprogramming. 🐸🧬
Posts
Media
Videos
Starter Packs
Pinned
Meghana Oak
@meghana-oak.bsky.social
· Jan 22

H3K4 methylation-promoted transcriptional memory ensures faithful zygotic genome activation and embryonic development
In the life of a vertebrate embryo, gene expression is initiated for the first time at zygotic genome activation (ZGA). Maternally expressed transcription factors present in the embryo are essential f...
doi.org
Reposted by Meghana Oak
Reposted by Meghana Oak
The Groth lab
@grothlab.bsky.social
· Feb 19
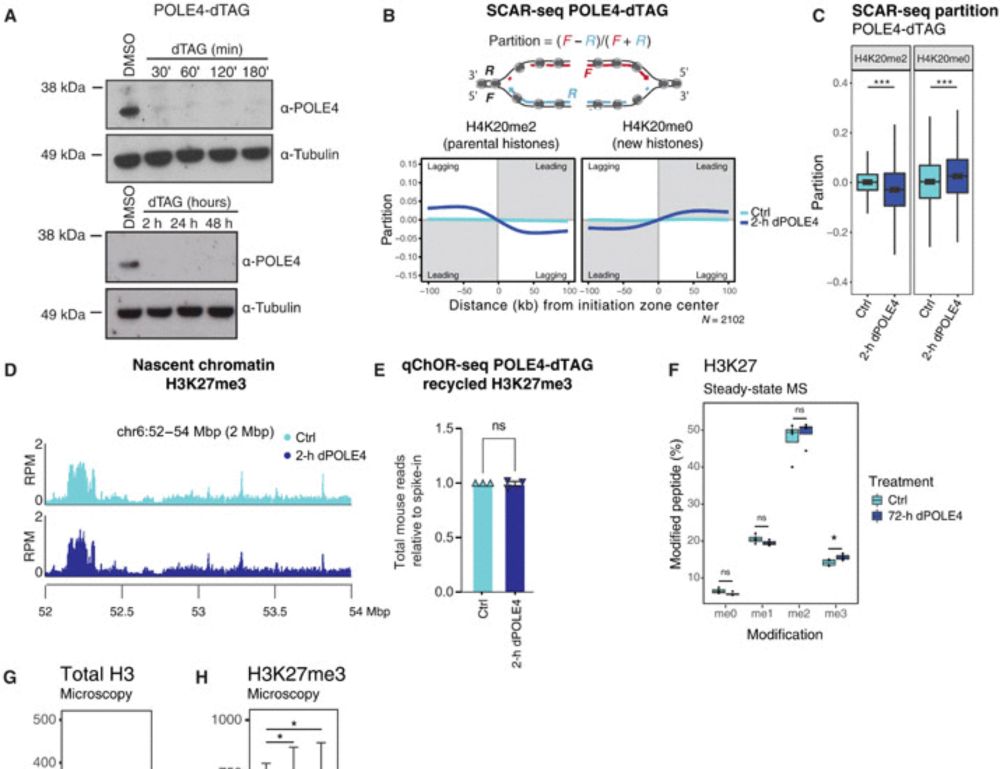
Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability
Losing parental histones during DNA replication fork passage challenges differentiation competence and cell viability.
tinyurl.com
Meghana Oak
@meghana-oak.bsky.social
· Feb 19
Reposted by Meghana Oak
Ana Janeva
@anajaneva.bsky.social
· Feb 6
Reposted by Meghana Oak
Ana Janeva
@anajaneva.bsky.social
· Jan 26

Digital Reprogramming Decodes Epigenetic Barriers of Cell Fate Changes
Fates of differentiated cells in our body can be induced to change by nuclear reprogramming. In this way, cells valuable for therapeutic purposes and disease modeling can be produced. However, the eff...
www.biorxiv.org
Meghana Oak
@meghana-oak.bsky.social
· Jan 22
Meghana Oak
@meghana-oak.bsky.social
· Jan 22
Meghana Oak
@meghana-oak.bsky.social
· Jan 22
Meghana Oak
@meghana-oak.bsky.social
· Jan 22
Meghana Oak
@meghana-oak.bsky.social
· Jan 22

H3K4 methylation-promoted transcriptional memory ensures faithful zygotic genome activation and embryonic development
In the life of a vertebrate embryo, gene expression is initiated for the first time at zygotic genome activation (ZGA). Maternally expressed transcription factors present in the embryo are essential f...
doi.org


